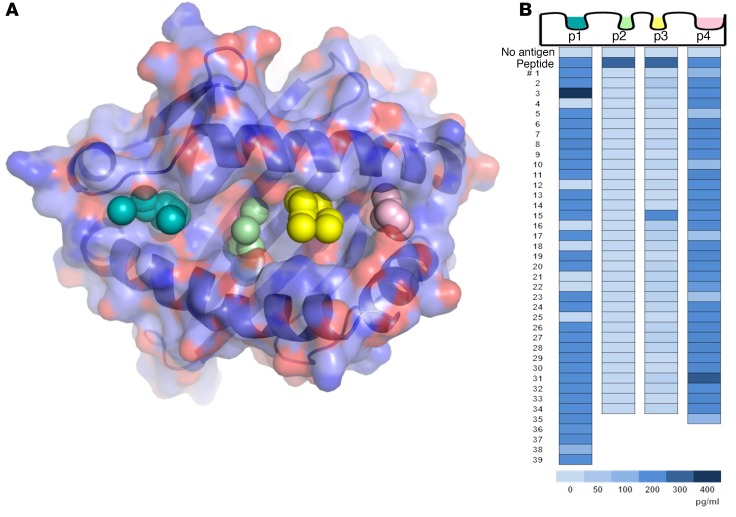
Figure 1. In silico screening of a small-molecule library by molecular docking to structural pockets within the HLA-DQ8 peptide–binding groove and in vitro screening of the top-scoring compounds.
(A) Molecular model of the DQ8 peptide binding cleft, with pockets 1, 4, 6, and 9 depicted by colored spheres. Using a supercomputer, 139,735 repository compounds were docked into each pocket in 1,000 different orientations and scored on the basis of the free energy of binding (ΔG). (B) The top-scoring compounds were screened with an in vitro bioassay, which used an immortalized T cell that responds to an insulin epitope presented by DQ8. The TCR α/β genes from an insulin B:9-23–restricted T cell clone were retrovirally transduced into a T cell line devoid of endogenous TCR to create a TCR transductant, named clone 5. Clone 5 was cultured with insulin peptide plus a small molecule in the presence of DQ8-Tg murine splenocytes. After overnight culture, secreted IL-2 was measured in the cell culture supernatant with a highly sensitive ELISA. In the heatmap, “No antigen” represents the response without in vitro–added peptide, and “Peptide” represents the response to peptide without an in vitro–added small molecule. Numbers 1–39 depict the tested compounds, and the compounds were different for each pocket. The insulin B:9-23 peptide was used to screen pockets 1 and 9 (p1 and p9), while insulin B:13-23 was used to screen pockets 4 and 6 (p4 and p6), which resulted in a greater baseline IL-2 response. Supplemental Tables 1–4 list the corresponding chemical structures, ΔG estimates from molecular docking, and rank based on the overall free energy score.