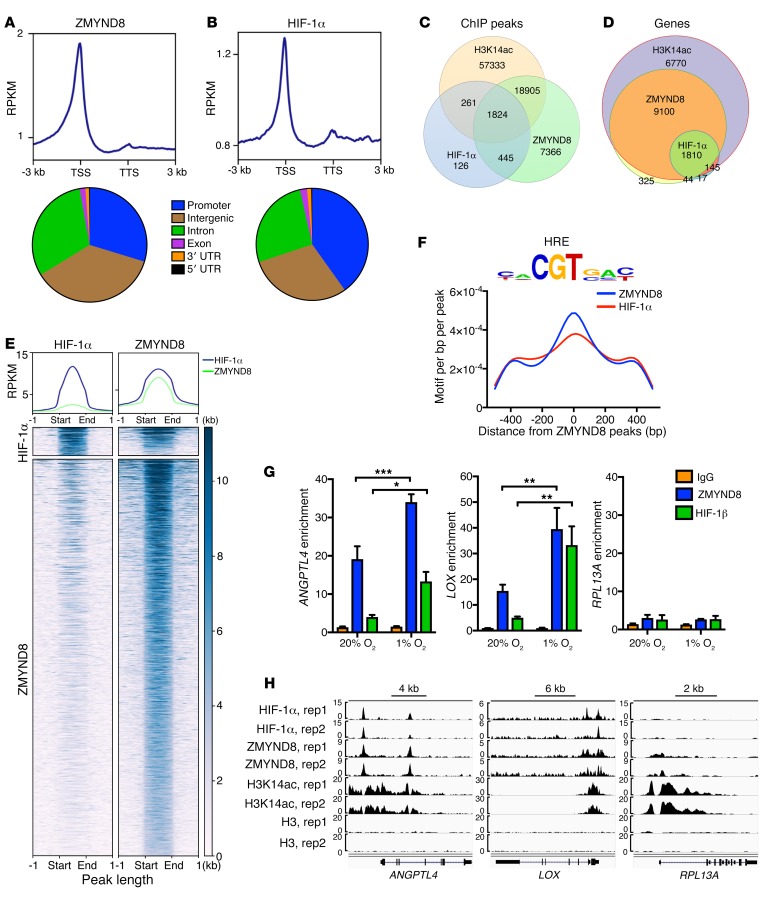
Figure 8. ZMYND8 globally colocalizes with HIF-1α at the HREs in breast cancer cells.
(A and B) Metagene analysis of the genomic distribution of ZMYND8 (A) and HIF-1α (B) in MDA-MB-231 cells exposed to 1% O2 for 24 hours (n = 2). RPKM, reads per kilobase per million mapped reads; TSS, transcription start site; TTS, transcription termination site. (C and D) Venn diagram of the overlapped ChIP-seq peaks (C) and co-occupied genes (D) by ZMYND8, HIF-1α, and H3K14ac (n = 2). (E) Co-occupancy analysis of HIF-1α and ZMYND8 ChIP-seq peaks (n = 2). (F) Motif density analysis of ZMYND8 ChIP-seq peaks (n = 2). HRE is shown in top panel. (G) ZMYND8 and HIF-1β ChIP-qPCR assays in MDA-MB-231 cells exposed to 20% or 1% O2 for 24 hours (mean ± SEM, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, by 2-way ANOVA with Sidak’s t test. (H) Genome browser snapshots of HIF-1α, ZMYND8, H3K14ac, and H3 ChIP-seq peaks. rep, replicate.