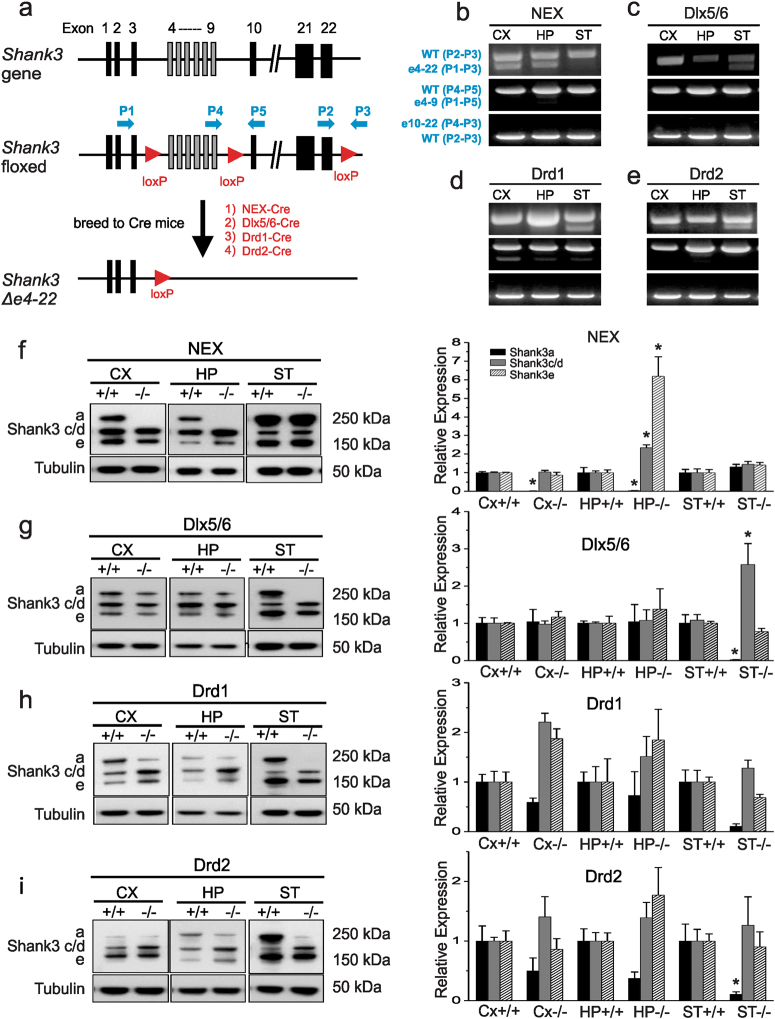
Fig. 1. Conditional Shank3 e4-22flox mice permit brain region-specific excision of Shank3.
a The wild-type mus Shank3 locus (top) depicting the engineered insertion of loxP sites (red arrowheads) before exon 4, after exon 9, and after exon 22 (middle). Crossing the Shank3 e4–22flox/flox mice to Cre mice results in a two-step recombination ultimately at the first and third loxP sites, yielding deletion of Δe4–22 in Cre-expressing cells (bottom). Primers (blue arrows) are shown for detecting recombination of the loxP sites. b–e PCR-based detection of Shank3 deletion of Δe4–22 (p1–p3), Δe4–9 (p1–p5), Δe10–22(p4–p3) in the cortex (CX), hippocampus (HP), and striatum (ST) of NEX-Cre Shank3 floxed mice (NEX) (b), Dlx5/6-Cre Shank3 floxed mice (Dlx5/6) (c), Drd1-Cre Shank3 floxed mice (Drd1) (d), and Drd2-Cre Shank3 floxed mice (Drd2) (e). A prominent deletion of e4–22 was observed in brain regions where corresponding Cres are predominantly expressed. f Western blots of dissected brains from NEX-Shank3 mice reveals a loss of Shank3a protein in CX and HP, but not in ST from crude PSD fractions (two-way ANOVA, main effects of genotype and region and interaction, p ≤ 0.0001); paradoxically, Shank3c/d and Shank3e were increased in the HP (two-way ANOVA, main effects of genotype and region and interaction, p ≤ 0.002); n = 5/region/genotype. g Western blotting of dissected brains from Dlx5/6-Shank3 mice reveals loss of Shank3a protein in the ST but not in the CX or HP crude PSD fractions (two-way ANOVA, main effects of region and interaction, p ≤ 0.05); with a similar paradoxical increase in Shank3c/d in the ST (two-way ANOVA, main effects of genotype and region and interaction, p ≤ 0.04) but no significant change for Shank3e; n = 5/region/genotype. h Western blotting of dissected brains of Drd1-Shank3 mice reveals a loss of Shank3a protein in the ST, but not in the CX or HP crude PSD fractions (two-way ANOVA, main effect of genotype, p-value ≤ 0.02), although this did not withstand Bonferroni-corrected post-hoc comparisons and no significant differences were seen for Shank3c/d or Shank3e; n = 5/region/genotype. i Western blotting of dissected brains of Drd2-Shank3 mice reveals loss of Shank3a protein in the ST but not in the CX or HP crude PSD fractions (two-way ANOVA, main effect of genotype, p-value ≤ 0.01), with no significant differences seen for Shank3c/d or Shank3e; n = 5/region/genotype. f–i, *p < 0.05, compared to the+/+control. All data are expressed as means ± SEM and were analyzed by two-way ANOVAs with genotype and brain region as factors; Bonferroni-corrected post-hoc comparisons