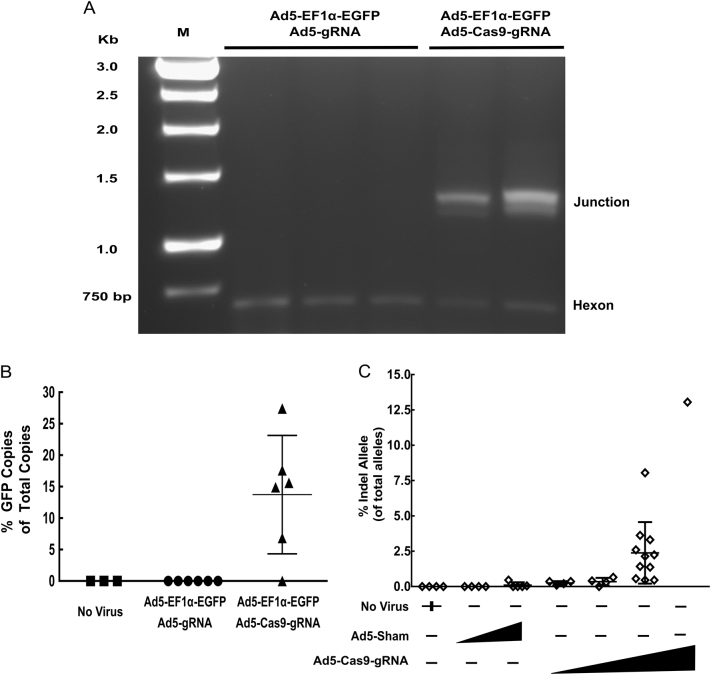
Fig. 5.
Knock-in and NHEJ rates following in vivo delivery with adenovirus is variable and persists over time. A total of 1 × 1011 viral particles of Ad5-EF1α-EGFP and Ad5-Cas9-gRNA (n = 6), or Ad5-EF1α-EGFP and sham Ad5-gRNA (n = 6), were injected into mice at a ratio of 1:1. PBS mock injected mice served as a no virus control (n = 3). a Junction capture PCR was used to amplify 3′ junctions from EGFP-integrated ROSA26 alleles from genomic DNA from the mice. Each lane represents genomic DNA template from one mouse. Viral amplification of 612 bp of the hexon gene served as a loading control and confirmed viral transduction. b LAM-PCR was performed by production of single-stranded linear PCR product from a biotinlyated primer binding downstream of the 3′ homology region of ROSA26 in the mouse genome, whereby both integrated and non-integrated alleles are amplified. qPCR analysis to quantify HDR-mediated integration rates is displayed as percent EGFP-integrated alleles out of total ROSA26 alleles. Error bars are s.d. of the mean. c Mice injected with Ad5-Cas9-gRNA or a sham virus were sacrificed at time points ranging between 7 and 210 days and genomic DNA was extracted. This DNA, and genomic DNA extracted at the conclusion of the hAAT experiment in Figs. 3 and 4, was used as template for targeted Illumina deep sequencing of the ROSA26 for the presence of edited alleles, witnessed by NHEJ-induced indels