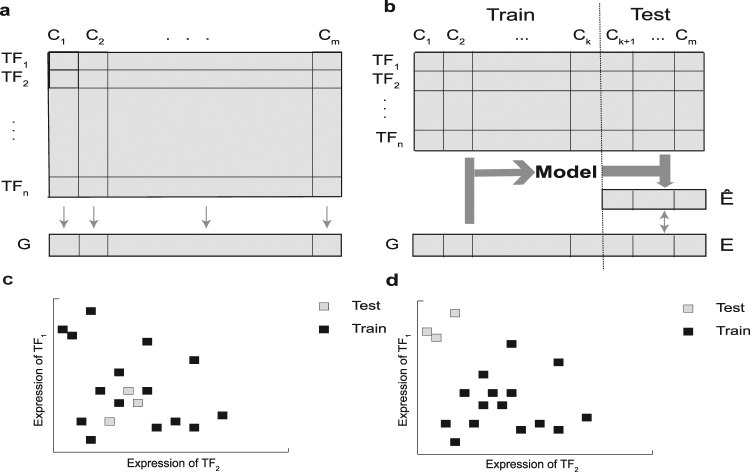
Figure 1.
Schematic illustration of expression-to-expression modeling, cross-validation and choice of test and training samples. (a) The goal is to predict expression of a gene G in conditions (i.e., samples) C1, C2, …, Cm using the expression of transcription factors TF1, TF2, …, TFn in the same conditions. (b) The schematic view of dividing conditions into training and test sets in cross-validation. A model is trained using the expression of TFs and gene G on the training set and is used to predict the gene expression in the test conditions. The predicted gene expression values (Ê) are compared to the real gene expression values (E) to provide a measure for the accuracy of the model. (c,d) The illustration of different ways of dividing conditions into test and training sets. In this toy example, only two TFs are considered and each point represents a condition. In (c) test points are located close to training points, but in (d) test points are far from training points. Evaluating the performance of a model such as the one shown in (a) using the partitions shown in (c) or (d) may lead to different results.