Figure 2.
LMN-1 Ser32 Phosphorylation Specifically Occurs at Meiotic Onset, and Is PLK-2- and CHK-2-Dependent and Independent of DNA DSBs
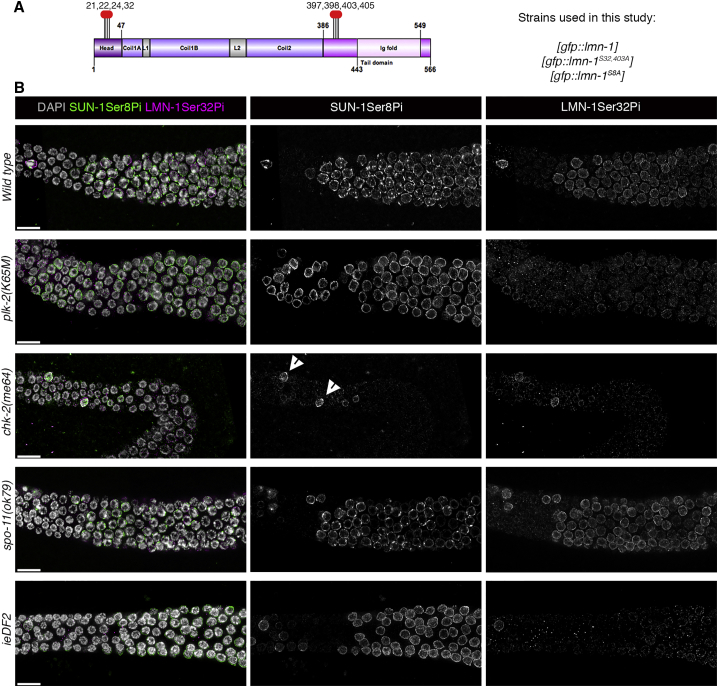
(A) Schematic representation of LMN-1, indicating phosphorylation sites (red circles) relevant to this study. Numbers indicate amino acid positions. Strains generated for this study express wild-type GFP::LMN-1 or GFP::LMN-1 containing point mutations altering serine/threonine residues to non-phosphorylatable alanine. In one mutant strain, serines 32 and 403 were changed to alanine; in the S8A mutant, Ser21, Ser22, Ser24, Ser32, Thr397, Ser398, Ser403, and Ser405 were changed to alanine residues (see Table S1 for viability counts). The schematic representation of the protein was designed using the software GPS-SUMO sp 2.0.
(B) Localization of LMN-1Ser32pi and SUN-1Ser8pi signals in the transition zone. The SUN-1Ser8pi signal specifically marks meiotic entry. Representative images are shown for the wild-type and plk-2, chk-2, spo-11, and ieDF2 mutants. Note the absence of meiotic phosphorylation at both SUN-1 Ser8 and LMN-1 Ser32 in the chk-2 mutant. Arrowheads indicate phospho-modified mitotic nuclei. Scale bars, 10 μm. Gonads shown in (B) consist of multiple maximum projection images stitched together to show larger sections of the gonads.
See Figure S2 for additional data.