Figure 4.
Mutants Expressing Non-phosphorylatable LMN-1 Have Delayed Leptotene Progression and Altered Heterochromatin Distribution in Transition Zone/Early Pachytene Nuclei
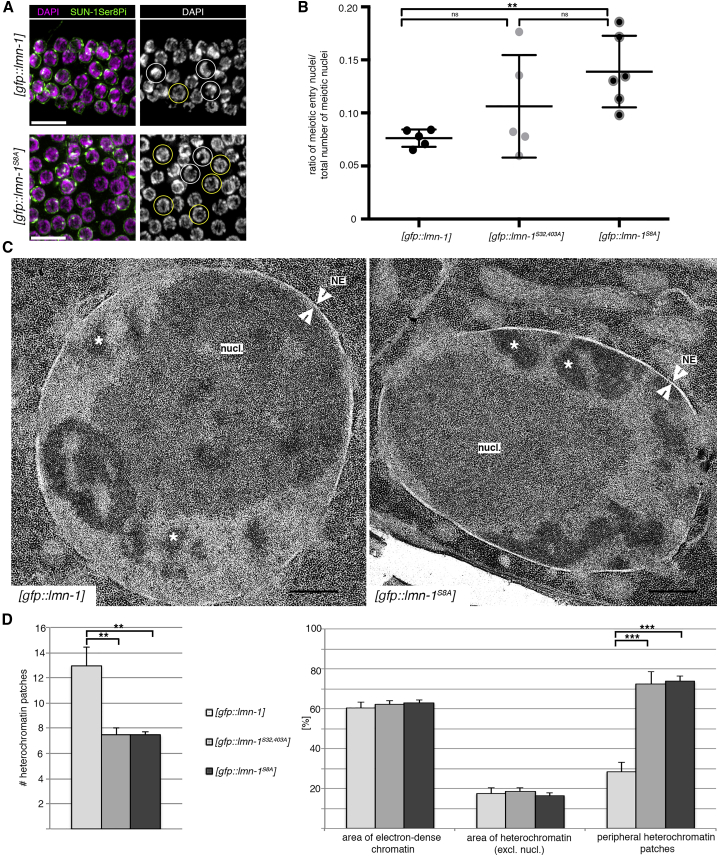
(A) Nuclei at meiotic entry have positive SUN-1Ser8pi staining and a non-polarized/spherical DAPI-staining pattern (yellow circles). Zygotene nuclei are also positive for SUN-1Ser8pi staining but have a polarized/crescent DAPI-staining pattern (white circles). Scale bars, 10 μm.
(B) Nuclei positive for SUN-1Ser8pi at meiotic entry in the wild-type and mutants expressing non-phosphorylatable LMN-1 were counted in all rows from meiotic entry until full SC polymerization, and values were normalized to the total number of nuclei from meiotic entry to diplotene. [gfp::lmn-1], n = 5 gonads; [gfp::lmn-1S32,403A], n = 5 gonads; [gfp::lmn-1S8A], n = 6 gonads. ns, not significant (p > 0.05); ∗∗p = 0.0043. p values were calculated using the Mann-Whitney U test. Scatter plots indicate the mean and SD. For additional classification of meiotic entry nuclei, see Figure S4.
(C) Representative electron micrographs of early meiotic nuclei from the wild-type and mutants expressing non-phosphorylatable LMN-1. Regions of higher electron density within the nucleus represent heterochromatin (star) and the large, polarized nucleolus (nucl.); arrowheads indicate the position of the nuclear envelope (NE). Scale bars, 0.5 μm.
(D) Number and distribution of heterochromatin patches in representative electron micrographs of nuclei from wild-type and mutants expressing non-phosphorylatable LMN-1. See Table S2 for further information on the choice of nuclei. [gfp::lmn-1], n = 9 nuclei; [gfp::lmn-1S32,403A], n = 8 nuclei; [gfp::lmn-1S8A], n = 8 nuclei. For the number of heterochromatin patches, ∗∗p = 0.005 for [gfp::lmn-1] versus [gfp::lmn-1S32,403A], ∗∗p = 0.004 for [gfp::lmn-1] versus [gfp::lmn-1S8A], and p > 0.05 (ns) is not indicated; for the percentage area of heterochromatin and peripheral heterochromatin, ∗∗∗p = 8.9 × 10−5 for [gfp::lmn-1] versus [gfp::lmn-1S32,304A], ∗∗∗p = 2.5 × 10−6 for [gfp::lmn-1] versus [gfp::lmn-1S8A], and p > 0.05 (ns) not indicated for all others. p values were calculated using the two-tailed Student's t test. Error bars represent SD.