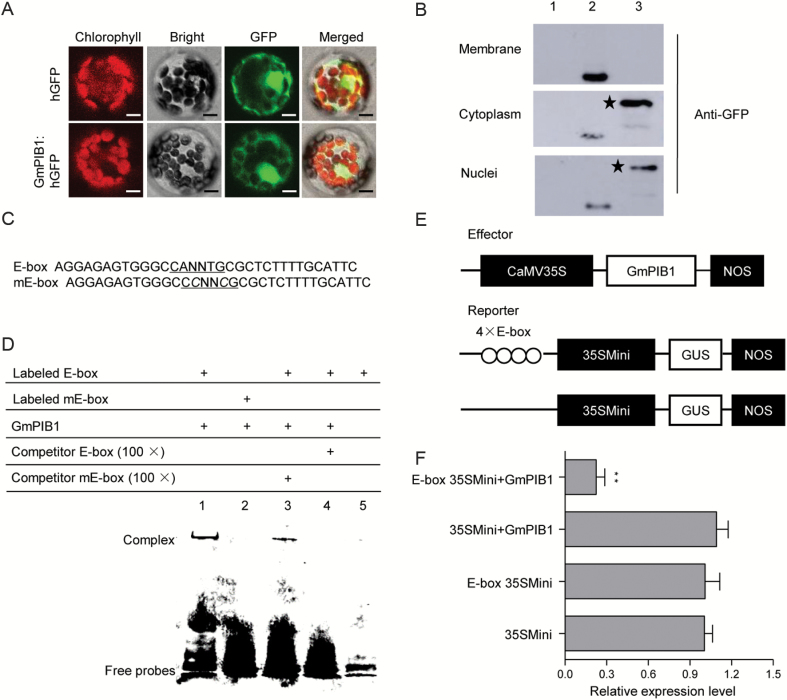
Fig. 3.
Sequence-specific binding activity of GmPIB1 to the E-box element. (A) Subcellular localization of GmPIB1–hGFP fusion protein. Subcellular localization was investigated in Arabidopsis protoplasts by confocal microscopy. The fluorescence from humanized GFP (hGFP) and the fusion protein GmPIB1–hGFP was observed under white light, UV light, and red light separately. Bars, 10 μm. (B) Immunoblot analysis detecting GmPIB1–hGFP fusion protein in the cytoplasm and nuclei. Line 1, Arabidopsis protoplasts (negative control); line 2, hGFP; line 3, GmPIB1–hGFP fusion protein. Anti-GFP was used to detect GmPIB1–GFP fusion protein in Arabidopsis cells. An asterisk denotes the specific band of the fusion protein GmPIB1–hGFP. (C) Nucleotide sequences of the E-box and mE-box probes. (D) EMSA showing sequence-specific binding of the recombinant GmPIB1 protein to the E-box. Lane 1, labeled E-box probe and GmPIB1 protein; lane 2, labeled mE-box probe and GmPIB1 protein; lane 3, titration using a cold mE-box sequence as a competitor; lane 4, titration using a cold E-box sequence as a competitor; lane 5, EMSA performed with only the free E-box probe. (E) Schematic diagram of the reporter and effector constructs. The reporter plasmids contained four repeats of the E-box sequence and 35Smini, and the effector plasmids encoded GmPIB1 under the control of the CaMV 35S promoter. (F) Relative GUS activity in transactivation assays. The effector and reporter plasmids were co-transfected into Arabidopsis protoplasts. The numbers show the fold increase in GUS activity compared with the vector E-box/35Smini promoter (E-box 35SMini) alone. The experiments were performed on three biological replicates and statistically analysed using Student’s t-test (**P<0.01). Bars indicate standard error of the mean. (This figure is available in color at JXB online.)