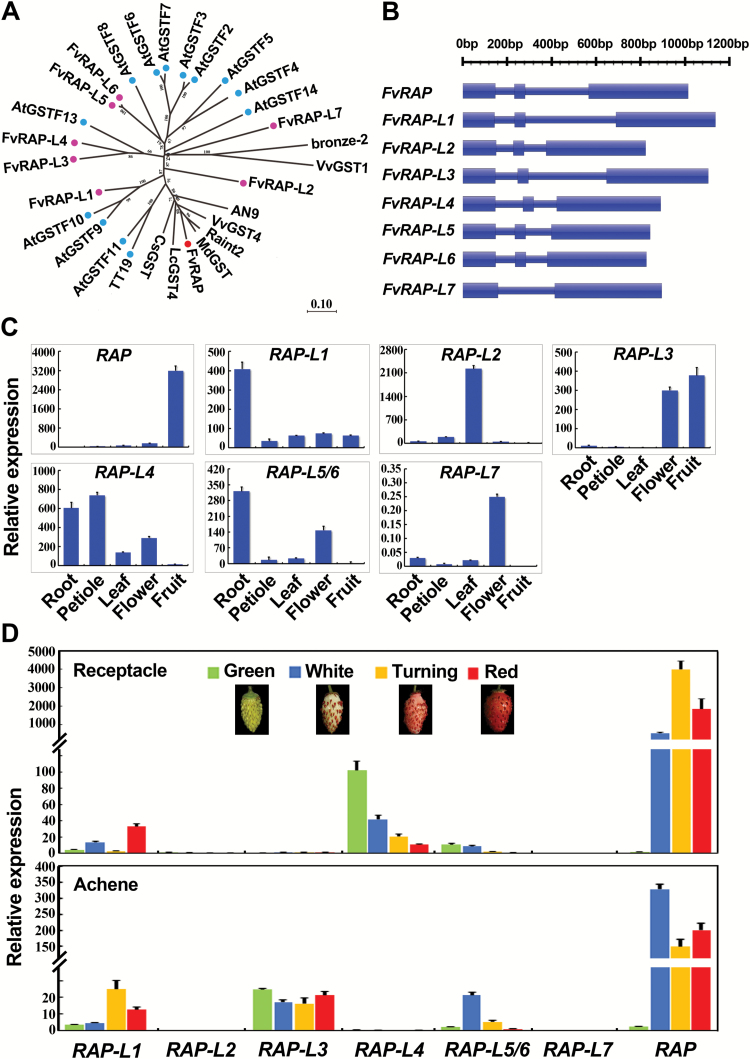
Fig. 2.
Phylogenetic analyses and expression patterns of RAP and its paralogs. (A) Phylogenetic tree of RAP and its homologs. A neighbor-joining tree was constructed based on protein sequences of RAP and its homologs from F. vesca, Arabidopsis, and several other species. Gene IDs are shown for genes from F. vesca and Arabidopsis, while the accession numbers in NCBI are shown for others: Riant2 (KT312848), MdGST (AEN84869), AN9 (Y07721), bronze-2 (AAV64226), VvGST4 (AAX81329), VvGST1 (AAN85826), LcGST4 (KT946768), CsGST (ABA42223), FvRAP (gene31672), FvRAP-L1 (gene28763), FvRAP-L2 (gene08595), FvRAP-L3 (gene22014), FvRAP-L4 (gene10549), FvRAP-L5 (gene10550), FvRAP-L6 (gene10551), FvRAP-L7 (gene10552), AtGSTF2 (At4G02520), AtGSTF3 (At2G02930), AtGSTF4 (At1G02950), AtGSTF5 (At1G02940), AtGSTF6 (At1G02930), AtGSTF7 (At1G02920), AtGSTF8 (At2G47730), AtGSTF9 (At2G30860), AtGSTF10 (At2G30870), AtGSTF11 (At3G03190), AtGSTF12/TT19 (At5G17220), AtGSTF13 (At3G62760), and AtGSTF14 (At1G49860). The numbers indicate the bootstrap values calculated from 1000 replicate analyses. (B) Gene models of RAP and its paralogs. Thick bars indicate exons, and thin bars indicate introns. The sequence length is shown at the top. (C) Expression patterns of RAP and its paralogs in the tissues of red-fruited Ruegen as analysed by qRT-PCR. (D) Expression patterns of RAP and its paralogs in fruit receptacles and achenes of Ruegen at four developmental stages, as analysed by qRT-PCR. Gene11892 was used as the internal control in (C) and (D). Data are means (±SD) obtained from three technical replicates. The experiment was repeated for three times with similar results.