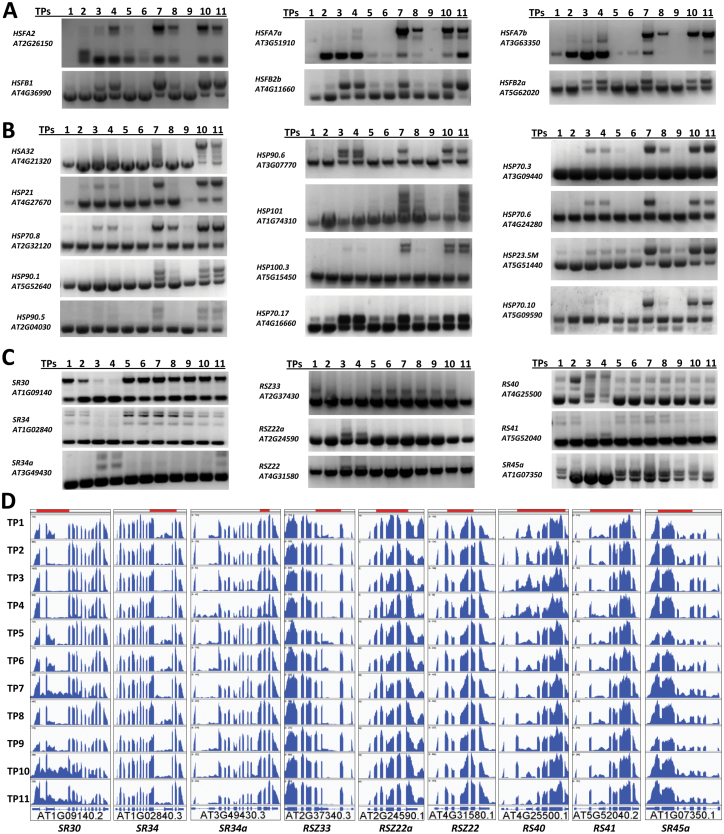
Fig. 8.
Differential splicing pattern of HSF genes, HSP genes and SR genes at different time-points. (A–C) RT-PCR was performed using primers that flank intron(s). (A) Splicing patterns of heat-shock factor (HSF) genes. HSFA2, HSFA7a, and HSFA7b expressed fewer intron-retained isoforms after heat priming (TP3 and TP4) when compared to after heat shock (TP7 and TP10). HSFB1, HSFB2a, and HSFB2b accumulated more intron-retained isoforms after heat priming when compared to after heat shock. (B) Splicing patterns of heat-shock protein (HSP) genes. genes show different ratios of intron retention after heat priming and heat shock. Most genes, except for HSP90.6 and HSP70.17, expressed fewer intron-retained isoforms after heat priming (TP3 and TP4) when compared to after heat shock (TP7 and TP10). (C) Splicing patterns of a group of serine-/arginine-rich (SR) genes. Specific of isoforms of SR genes were induced in heat priming (TP3 and TP4). (D) Gene structure and intron retention of interesting regions from the nine SR genes in (C) as illustrated using the IGV program (http://software.broadinstitute.org/software/igv/). Coverage of RT-PCR-amplified fragments are indicated by the shaded bars at the top of each panel. (This figure is available in colour at JXB online.)