Abstract
Microvesicles (MVs) are secreted by multiple types of tumor cell and are involved in tumor progression and metastasis. The aim of the present study was to explore the effects of MVs derived from salivary adenoid cystic carcinoma (SACC) and to investigate their potential involvement in the pathogenesis of perineural invasion of SACC. MVs were isolated from ACCs cells, and differential gene expression profiles of these MVs were compared with their donor cells to speculate on their biological functions. Several candidate genes were validated using reverse transcription-quantitative polymerase chain reaction analysis. The effects of ACCs MVs on rat Schwann cells (RSC96 cells), which are the principal glia of the peripheral nervous system, were then evaluated by phospho-antibody array performed on RSC96 cells transduced with ACCs MVs. The results indicated that ACCs cells may produce MVs. Microarray-based expression profiles between ACCs cells and their MVs identified 1,355 genes involved in cell adhesion, development and the regulation of apoptosis. In addition, the extracellular signal-regulated protein kinase signal pathway in RSC96 cells may be induced by ACCs-derived MVs. These results may help to elucidate the mechanisms underlying perineural invasion in SACC, and to determine a promising anti-tumor biological therapeutic target.
Keywords: microvesicle, adenoid cystic carcinoma, microarray, protein array, Schwann cell, extracellular signal-regulated kinase
Introduction
Salivary adenoid cystic carcinoma (SACC) is a malignant tumor of the salivary gland, with a poor prognosis and unknown etiology (1). Perineural invasion, which is a distinctive biological characteristic of SACC, leads to permanent regional dysfunction, local tumor recurrence and shorter survival time in contrast with other salivary tumors, despite definitive treatment (2). Thus, it is important to unveil the mechanisms underlying perineural invasion to develop more effective therapeutic strategies. SACC cells and their neighbouring nerve tissues have previously been demonstrated to interact through the release of several molecular substances, which may alter the surrounding microenvironment of nerve tissues to facilitate tumor cell gowth, proliferation and invasion (3).
Microvesicles (MVs) are spherical or saucer-shaped, bilayer lipid membrane-bound vesicles, which are heterogeneous in size (range, 30–1,000 nm) (4,5). MVs originate from multiple types of tumor cell, as well as hematopoietic and epithelial cells, under normal and pathological conditions (6–8). Two cellular vesiculation mechanisms have been identified. The first is an inward budding of endosomal membranes, creating multivesicular bodys that later fuse with the plasma membrane, releasing the intraluminal vesicles as exosomes (30–100 nm in diameter) into the extracellular environment (9,10). The second is the direct outward blebbing of the cellular plasma membrane, termed shedding vesicles or ectosomes (100–1,000 nm in diameter) (9,10). MV cargos have previously been confirmed to include a variety of lipids, proteins and nucleic acids (11). The composition and biological function of the MVs depend on the donor cells (12–14) For instance, T cell-derived MVs may be involved in the antigen presentation procedure (14), and tumor-derived MVs may suppress the immune response and promote tumor growth (13,14). Growing evidence has indicated that MVs act as mediators that deliver abundant bioactive molecules involved in intracellular communication, in contrast with traditional cell-to-cell contact-dependent signaling exchange (15–17).
To the best of our knowledge, no studies exist in the literature concerning MVs derived from SACC cells. In the present study, it was hypothesized that MVs sercreted by SACC cells were associated with communication between tumor cells and the neighboring nerve tissues. First, MVs derived from the human SACC cell lines, ACC-2, were described, and differential gene expression profiles of these MVs were established and compared with those of their donor cells to speculate on their biological functions. Several candidate genes were also validated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. The effects of ACC-2 MVs on RSC96 rat Schwann cells, which are the principal glia of the peripheral nervous system, were then evaluated by a phospho-antibody array performed on RSC96 cells transduced with ACC-2 MVs. The results of the present study may help to elucidate the mechanisms underlying perineural invasion in SACC, and to determine a promising anti-tumor therapeutic target.
Materials and methods
Cells and reagents
The human salivary adenoid cystic carcinoma ACC-2 cell line and rat RSC96 Schwann cells were obtained from the State Key Laboratory of Oral Diseases, Sichuan University (Chengdu, China) and the American Type Culture Collection (Manassas, VA, USA), respectively. Primary antibodies and rabbit anti-mouse horseradish peroxidase-conjugated secondary antibodies for human major histocompatibility complex (MHC) class I (cat. no., sc71256; dilution, 1:100), heat shock protein 70 (HSP70; cat. no., sc-24; dilution, 1:100) and β-actin (cat. no., sc-130065; dilution, 1:1,000) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Cell culture and scanning electron microscopy
The ACC-2 cell line was cultured in vitro at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% MV-free fetal bovine serum (dFBS; Gibco; Thermo Fisher Scientific, Inc; prepared by ultracentrifugation at 110,000 × g at 4°C for 16 h to remove bovine MVs), 100 U/ml penicillin G and 100 mg/ml streptomycin. For scanning electron microscopy (SEM), the cells were grown on coverslips, fixed with 2.5% glutaraldehyde at 4°C for 24 h and dehydrated in a series of increasing ethanol concentrations (30–100%). The cells were then transferred to a Hitachi HCP-2 Critical Point Dryer (Hitachi High-Technologies Corporation, Tokyo, Japan) followed by covering with gold, and observed using the Hitachi S3400 scanning electron microscope (Hitachi High-Technologies Corporation).
MV isolation
ACC-2-derived MVs (ACC-2 MV) were collected from conditioned medium by differential centrifugation as described below, with a number of modifications. Briefly, the supernatant was harvested following culture for 48 h, centrifuged at 4°C sequentially at 300 × g for 10 min, 2,000 × g for 20 min and 16,500 × g for 30 min to eliminate floating cells, debris and large membrane fragments, followed by filtration through a 0.22 µm filter. MVs were pelleted by ultracentrifugation at 110,000 × g at 4°C for 70 min using a SW41 rotor (Beckman optima L-80XP; Beckman Coulter, Inc., Brea, CA, USA). The pellet was washed twice in PBS, centrifuged at 110,000 × g at 4°C for 70 min to pellet again, and then resuspended in PBS and stored the solution at −80°C until use. Total protein content of the MVs was measured using a bicinchoninic acid protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer's protocol.
Transmission electron microscopy (TEM)
TEM was performed in the Electron Microscopy office of the College of Basic and Forensic Medicine, Sichuan University (Chengdu, China). ACC-2 MVs (20 µl) were loaded onto formvar carbon-coated grids at room temperature for 1 min without fixing, stained with a drop (20 µl) of 1% phosphotungstic acid for 1 min at room temperature, dried at room temperature for 10 min and examined using a Hitachi H600-4 electron microscope (Hitachi High-Technologies Corporation).
Western blot analysis
Total proteins were extracted using a total protein extraction kit (Jiangsu KeyGen BioTech Co., Ltd., Nanjing, China) according to the manufacturer's protocol. Samples were then lysed on ice for 30 min, then centrifuged at 12,000 × g at 4°C for 15 min. A total of 20 µl ACC-2 microvesicular or cell-lysate proteins were subjected to 10% SDS-PAGE. Following transfer to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA) and blocking with 5% non-fat milk for 1 h at room temperature, successive incubations with MHC class I, HSP70 and β-actin antibodies were performed overnight at 4°C, and with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Immunoreactive proteins were then detected using an enhanced chemiluminescence system (Bio-Rad Laboratoires, Inc.). The bands were scanned using a densitometer (Bio-Rad Laboratories, Inc.), and quantification was performed using Quantity One v.4.6.3 software (Bio-Rad Laboratories, Inc.).
RNA extraction and purification
Total RNA from ACC-2 MVs (400 µg/ml) or the donor cells (1 µg/ml; 1×106) was extracted and purified using an RNeasy mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol, and checked for a registrant identification number to inspect RNA integration using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA).
Microarray analysis
The microarray experiments were performed by Shanghai Biotechnology Co., Ltd. (SBC; Shanghai, China) using the 4×44K Whole Human Genome Oligo Microarray (Agilent Technologies, Inc.). The array was performed on two different RNA samples (ACC-2 cells or ACC-2 MV). In brief, the amplification and labeling of 500 ng of total RNA was performed according to the Agilent Low Input Quick Amp Labeling kit's protocol using Cy3 (Agilent Technologies, Inc.). Each slide was hybridized with 1.65 µg Cy3-labeled cRNA using a Gene Expression Hybridization kit (Agilent Technologies, Inc.) for 17 h at 50°C, according to the manufacturer's protocol. Following hybridization and washing with buffer (0.1X saline sodium citrate, 0.5% SDS), the processed slides were scanned with the Agilent Microarray Scanner (Agilent Technologies, Inc.). The resulting text files were extracted from Agilent Feature Extraction Software (version 10.7; Agilent Technologies, Inc.), and the raw data were normalized by a Quantile algorithm using the Agilent Gene Spring software (version 11.0; Agilent Technologies, Inc.). To identify the differentially expressed genes, a fold-change screening between the two samples obtained from the experiment was performed. The threshold used to screen up- or downregulated genes was a fold change of >2. The SBC Analysis System (SAS, Shanghai Biotechnology Co., Ltd.), a web-based program for conducting the subsequent data analysis and generating the results table, is available at http://sas.ebioservice.com/. A Gene Ontology database search (http://www.geneontology.org/) was performed to identify the functions of differentially-expressed mRNA transcripts between MVs and cells.
RT-quantiative PCR (RT-qPCR)
A total of 500 ng RNA (from ACC-2 cells or ACC-2 MVs) was subsequently reverse transcribed to cDNA using a Superscript II reverse transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protcol. RT-qPCR was performed using the SYBR® premix Ex Taq™ II kit (Takara Bio, Inc., Otsu, Japan) according to the following two-step thermocycler protocol: 1 cycle at 95°C for 30 sec, and then 40 cycles at 95°C for 5 sec, then 60°C for 31 sec. The following PCR primers were used: β-actin forward, 5′-ATGGATGACGATATCGCTGC-3′ and reverse, 5′-CACACTGTGCCCATCTACGA-3′; platelet-derived growth factor receptor α (PDGFRA) forward, 5′-CTCTCCCTTGTACAGCCTTATTTTG-3′ and reverse, 5′-TCCGGCCTCATGATGTCA-3′; fibroblastic growth factor 12 (FGF12) forward, 5′-GGAAAAGGATGCTAGATGCTGA-3′ and reverse, 5′-TTTAACTTGGGCTCCAGGAA-3′; HLA class II histocompatibility antigen, DO β chain (HLA-DOB) forward, 5′-TGCCTGGGCGTGTTGAA-3′ and reverse, 5′-CCAGCCTACTGCAGGTTGTTT-3′; and interleukin-19 (IL-19) forward, 5′-ACGCTGCTGCCATTAAATCC-3′ and reverse, 5′-CATCAAGCTGAGAACATTACTTCATG-3′. The comparative threshold cycle method was used to calculate amplification fold. β-actin was used as a reference control gene to normalize the expression value of target genes. Triple replicates were performed for each gene, and the average expression value was computed for subsequent analysis. The relative expression level of the genes was calculated using the 2−ΔΔCq method (18).
Internalization of MVs by Schwann cells
MVs (5 µg) derived from ACC-2 cells were labeled with a DiI red fluorescent probe (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol. The labeling step was stopped by washing three times in PBS, and the ACC-2 MVs were then pelleted by ultracentrifugation at 110,000 × g for 70 min at 4°C. MV-free supernatant was used as a control to determine the effects of any free fluorescent dye present in the PBS used for suspension of the MVs. The labelled ACC-2 MVs or the MV-free supernatant were mixed with 1×104 RSC96 cells (rat Schwann cells) and incubated in DMEM containing 10% FBS. MVs were allowed to bind for 30 min at 37°C and the binding was then stopped by washing twice in cold PBS. A Leica DM16000B fluorescence microscope at magnification, ×100 (Leica Microsystems GmbH, Wetzlar, Germany) was used for observation.
Phospho-protein profiling by phospho-antibody array
RSC96 cells were cultured in DMEM containing 10% dFBS only, or supplemented with ACC-2 MVs (50 µg/1×106 cells) in the described medium for 24 h at 37°C with 5% CO2. Cell lysates obtained from RSC96 or ACC-2 MV-transduced RSC96 cells were applied to the Extracellular Signal-Regulated Kinase (ERK) Signaling Phosphorylation Antibody Array, which was purchased from and performed by Full Moon BioSystems, Inc. (Sunnyvale, CA, USA). The array contained 202 antibodies, each of which had 6 replicates, on glass microscope slides. Briefly, 100 µg cell lysate in 50 µl reaction mixture was labeled with 1.43 µl biotin in 10 µg/µl of N,N-dimethyformamide. The resulting biotin-labeled proteins were diluted to a ratio of 1:20 in coupling solution prior to applying them to the array for conjugation. To prepare the antibody microarray, it was first blocked with blocking solution for 30 min at room temperature, rinsed with Milli-Q grade water (EMD Millipore, Billerica, MA, USA) for 3 min, and then dried with compressed nitrogen. Finally, the array was incubated with biotin-labelled cell lysates at 4°C overnight. The array slide was subsequently washed three times with 60 ml 1X wash solution for 10 min each, and the biotin-labeled protein was detected using Cy3-streptavidin. The slides were scanned on a GenePix 4000 scanner (Molecular Devices, LLC, Sunnyvale, CA, USA) and the images were analyzed with GenePix Pro 6.0 software (Molecular Devices, LLC). For each antibody, the following phosphorylation ratio was computed (phosphorylated and matching unphosphorylated values are denoted by phospho and unphospho in the control data and experiment data):
PANDA software (version 2.0; available from http://www.mathcs.emory.edu/panda/index.html) was utilised to allow the quantitative evaluation of the change in phosphorylation at each site with a 95% confidence interval.
Statistical analysis
All results were expressed as the mean ± standard deviation, when normally distributed. The statistical significance of differences was assessed using the Student's two-tailed t-test in two groups and one-way analysis of varience in multiple groups. The Student-Newman-Keuls method was used for post-hoc analysis. P<0.05 was considered to indicate a statistically significant difference. All data were analyzed with SPSS 15.0 software (SPSS, Inc., Chicago, IL, USA).
Results
MVs derived from ACC-2 cells
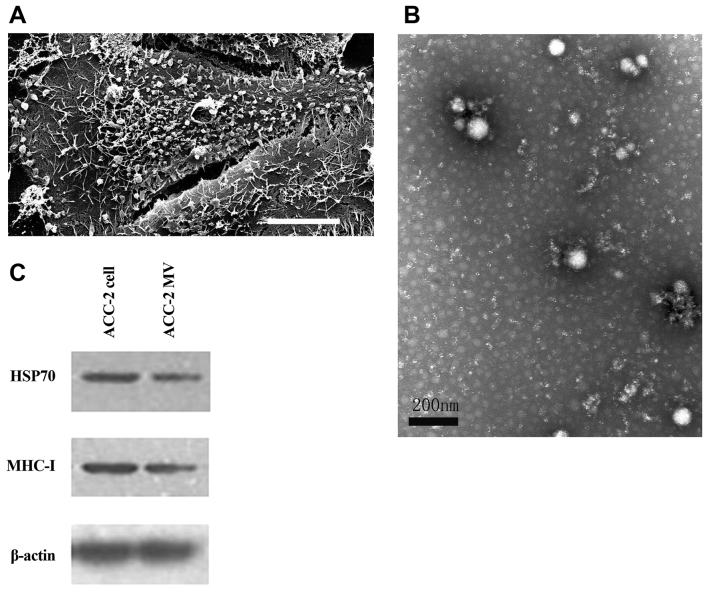
The ACC-2 cells were maintained in vitro at 37°C in 5% CO2 in complete medium. To obtain purified ACC-2 cell-derived MVs, the FBS added to the medium should be depleted of MVs (dFBS). To determine whether the ACC-2 cells produced MVs, SEM was performed on these cells following 48 h of culture. The SEM images revealed that the ACC-2 cells were covered with MV-like structures varying in size from 30–500 nm (Fig. 1A). MVs were prepared by differential centrifugation from medium supernatants as previously described (19). To confirm that the structures studied were MVs, the pellets were examined by TEM for morphological detection and western blot analysis of molecular markers. The transmission electron micrographs of the ACC-2 MVs revealed rounded structures with a size of 30–100 nm, similar to previously described MVs (Fig. 1B) (4). The identity of the studied vesicles was confirmed as MVs by western blot analysis (Fig. 1C), which revealed the presence of the proteins MHC-I and HSP70, which are commonly used markers for MVs (14,20). Therefore, ACC-2 cells may produce MVs, and MVs were isolated from ACC-2 cells for subsequent studies.
Figure 1.
Characterization and identification of ACC-2 cell-derived MVs. (A) The scanning electron micrograph revealed that an ACC-2 cell was covered with MV-like structures varying in size from 30–500 nm. Scale bar, 10 µm. (B) ACC-2 MVs were isolated by differential centrifugations from medium supernatants and the pellets were examined by TEM. TEM image of the ACC-2 MVs depicting rounded structures with a size of 30–100 nm. Scale bar, 200 nm. (C) HSP70 and MHC-I protein levels in ACC-2 cells and their microvesicles were detected using western blot analysis. The β-actin loading control is also shown (n=3). MV, microvesicle; TEM, transmission electron microscopy; MHC-I, major histocompatibility complex class I; HSP70, heat shock protein 70.
Gene expression profile analysis
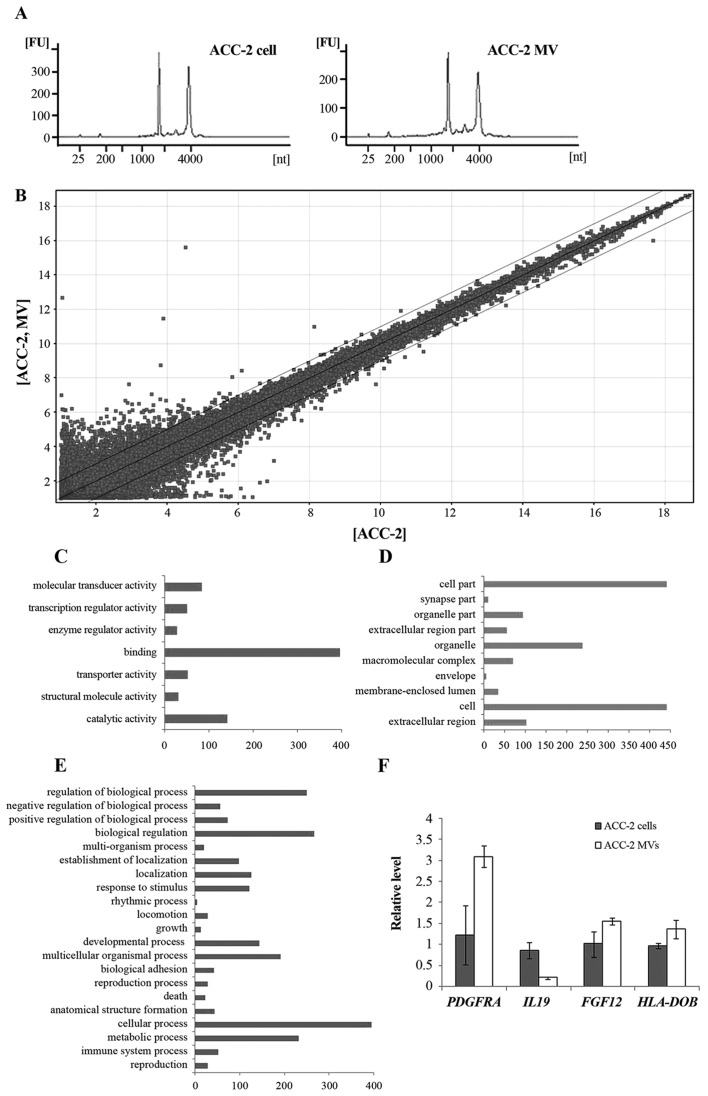
In the present study, bioanalysis of the RNA extracted from ACC-2 MVs demonstrated that ACC-2 MVs contained a broad range of RNA sizes, consistent with those characteristic of cellular RNA, however in a previous study, 18S and 28S ribosomal RNA peaks were absent in other types of MVs (Fig. 2A) (21). Microarray analysis of mRNA populations in MVs and their donor ACC-2 cells was performed using the Agilent 44K whole genome microarray. Following processing of expression data, 29,352 gene transcripts were identified in the cells and 29,298 transcripts were identified in the MVs (detected at levels above background) on the arrays. A total of ~900 different mRNAs were detected exclusively in MVs, indicating a selective enrichment process within the MVs. A total of 1,355 transcripts were revealed to be differentially distributed >2-fold (Fig. 2B). Of these, 641 transcripts were overexpressed (≤3,220-fold) and 714 transcripts were underexpressed in MVs compared with donor cells (up to 40-fold). Gene functional annotation using Gene Ontology was performed to identify the differentially-expressed mRNA transcripts between MVs and cells (Fig. 2C-E). These genes were revealed to be associated with cell adhesion, cytoskeleton, cell defense, cell metabolism, development, cell cycle and signal transduction.
Figure 2.
Characterization of the ACC-2 MV RNA. (A) Bioanalyzer data revealed the size distribution of total RNA extracted from ACC-2 cells and their MVs. ACC-2 MV contained a broad range of RNA sizes, consistent with the characteristics of cellular RNA. (B) A scatter plot of differentially expressed mRNA transcripts between MVs and cells. GO annotation by (C) molecular function, (D) cellular component and (E) biological process. (F) Quantitative analysis of transcript levels of 4 genes identified by microarray analysis, relative to β-actin, determined by reverse transcription-quantitative polymerase chain reaction. Error bars indicate the mean ± standard deviation (n=3). MV, microvesicle; GO, Gene Ontology; PDGFRA, platelet-derived growth factor receptor α; IL19, interleukin-19; FGF12, fibroblastic growth factor 12; HLA-DOB, HLA class II histocompatibility antigen, DO β chain.
RT-qPCR validation of differentially-expressed genes
There were multiple differentially-expressed genes in the gene chips. In the present study, the expression of four genes (IL-19, PDFGRA, HLA-DOB and FGF12) was also validated by RT-qPCR (Fig. 2F). To obtain truly comparable results, unamplified total RNA (from the same batch used for array hybridizations) was used as the template. The RT-qPCR results revealed that the expression levels of the four genes were consistent with that from the microarray analysis.
Internalization of MVs by Schwann cells
To determine whether the MVs contenting bioactive molecules were internalized by recipient cells, the ACC-2 MVs labelled with DiI red fluorescent dye or the MV-free supernatant were incubated with RSC96 cells (rat Schwann cells) in culture. Red fluorescent-positive cells were observed, indicating that the MVs effectively fused with the RSC96 cell membrane and entered the RSC96 cells (Fig. 3), similar to the results of a previous study (22).
Figure 3.
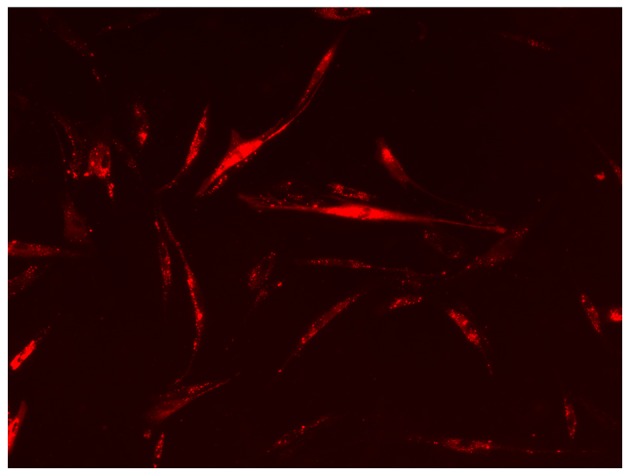
ACC-2 MVs are internalized into RSC96 cells. The ACC-2 MVs labelled with the membrane dye DiI (red) or MV-free supernatant were added to RSC96 cells in culture. The cells were then observed using a fluorescence microscope at magnification, ×100. The image depicts red fluorescent-positive RSC96 cells. MV, microvesicle.
Phospho-protein profiling by phospho-antibody array
To determine whether ACC-2 MVs affect Schwann cells, a phospho-specific antibody microarray for the ERK signaling pathway was used to compare the phosphorylation status of direct and indirect ERK targets in ACC-2 MV-transduced RSC96 cells and untreated RSC96 cells. This antibody array included 202 specific and well-characterized phosphospecific antibodies for proteins in the ERK pathway, each with six replicates. The paired antibodies for the same (but unphosphorylated) target sites were also included in the array to allow determination of the relative level of phosphorylation. Based on these features, a PANDA software was utilised to allow the quantitative evaluation of the change in phosphorylation at each site with a 95% confidence interval. A spectrum of proteins was identified, where phosphorylation levels were increased or decreased by >20%, with low values of 95% confidence invterval in RSC96 cells induced with ACC-2 MVs. Using a cutoff ratio of 0.8, 15 sites were identified that were hypophosphorylated in ACC-2 MV-transduced RSC96 cells compared with untreated RSC96 cells (Table I). Using a cutoff ratio of >1.2, 31 sites were identified with increased phosphorylation (Table II). These proteins were associated with cell growth, angiogenesis, cell defferentiation, inhibitation of apoptosis, development of nervous system and signal transduction. A number of these proteins were transcription factors that, when phosphorylated, modulate the expression of target genes. These transcription factors included cAMP responsive element binding protein, ETA domain-containing protein, ribosomal protein S6 kinase A5 and signal transducer and activator of transcription 3. Furthermore, the hyperphosphorylation of several sites, including phospholipase C, protein-tyrosine kinase 2-β, protein kinase C and growth factor receptor-bound protein 2, indicated that ACC-2 MVs may contain multiple types of growth factors. Thus, ACC-2 MVs appear to affect Schwann cell lines through activation of the ERK signaling pathway.
Table I.
Increase in phosphorylation of ERK signaling pathway components following incubation of RSC96 cells with ACC-2 MV.
| RSC96 | RSC96+MV | RSC96+MV/RSC96 | ||||
|---|---|---|---|---|---|---|
| Phosphorylation site | Ratio | 95% CI | Ratio | 95% CI | Ratio | Biological effects |
| BAD (phospho-Ser134) | 0.45 | 0.43–0.46 | 0.59 | 0.56–0.62 | 1.32 | Apoptosis, inhibited |
| BAD (phospho-Ser136) | 0.94 | 0.91–0.97 | 1.3 | 1.28–1.32 | 1.37 | Apoptosis, inhibited |
| B-RAF (phospho-Ser446) | 1.11 | 1.07–1.15 | 1.36 | 1.26–1.47 | 1.23 | Cell growth, altered |
| B-RAF (phospho-Ser601) | 1.07 | 1.05–1.09 | 1.38 | 1.32–1.44 | 1.30 | Signal transduction |
| CaMK1-α (phospho-Thr177) | 0.26 | 0.23–0.29 | 0.53 | 0.48–0.58 | 2.07 | Signal transduction |
| CaMK2-β/γ/δ (phospho-Thr287) | 0.82 | 0.8–0.85 | 1.23 | 1.18–1.27 | 1.49 | Nervous system development |
| CaMKII (phospho-Thr286) | 0.93 | 0.89–0.96 | 1.16 | 1.02–1.3 | 1.25 | Cell growth, altered |
| c-Raf (phospho-Ser43) | 0.63 | 0.57–0.69 | 0.82 | 0.78–0.86 | 1.29 | Cell growth, altered |
| CREB (phospho-Ser129) | 1.35 | 1.33–1.37 | 2.37 | 2.12–2.62 | 1.75 | Cell growth, altered |
| CREB (phospho-Ser133) | 1.24 | 1.21–1.28 | 1.52 | 1.5–1.53 | 1.22 | Activation |
| Elk1 (phospho-Thr417) | 1.82 | 1.77–1.86 | 10.17 | 9.39–10.95 | 5.53 | Transcription, DNA-dependent |
| ERK3 (phospho-Ser189) | 1.7 | 1.65–1.75 | 2.91 | 2.81-3 | 1.72 | Angiogenesis |
| FosB (phospho-Ser27) | 1.38 | 1.3–1.45 | 2.21 | 1.98–2.44 | 1.54 | Transcription, altered |
| GRB2 (phospho-Ser159) | 0.91 | 0.9–0.92 | 1.3 | 1.27–1.34 | 1.41 | Cell differentiation |
| MKK3 (phospho-Ser189) | 0.47 | 0.45–0.49 | 0.89 | 0.81–0.97 | 1.91 | Angiogenesis |
| MKK7/MAP2K7 (phospho-Ser271) | 0.5 | 0.46–0.53 | 1 | 0.84–1.15 | 2.02 | Transcription, altered |
| MSK1 (phospho-Ser360) | 0.55 | 0.48–0.62 | 0.75 | 0.69–0.82 | 1.41 | Enzymatic activity, induced |
| PAK2 (phospho-Ser192) | 1.57 | 1.51–1.63 | 1.97 | 1.86–2.07 | 1.27 | Regulation of growth |
| PAK3 (phospho-Ser154) | 1.68 | 1.63–1.73 | 2.05 | 1.97–2.13 | 1.22 | Axonogenesis |
| PKCδ (phospho-Thr505) | 0.67 | 0.65–0.7 | 0.89 | 0.87–0.91 | 1.32 | Apoptosis, altered |
| PKCθ (phospho-Ser676) | 1.68 | 1.57–1.78 | 2.15 | 1.87–2.43 | 1.28 | Apoptosis, altered |
| PKR (phospho-Thr446) | 1.46 | 1.41–1.51 | 3.93 | 3.78–4.08 | 2.69 | Cell cycle regulation |
| PKR (phospho-Thr451) | 1.17 | 1.14–1.19 | 1.68 | 1.56–1.79 | 1.43 | Enzymatic activity, induced |
| PLCβ3 (phospho-Ser537) | 1.05 | 1.02–1.08 | 1.35 | 1.3–1.4 | 1.28 | G-protein coupled receptor Protein signaling pathway |
| PLCG1 (phospho-Tyr771) | 0.74 | 0.68–0.8 | 1.06 | 1.01–1.11 | 1.43 | Cell cycle regulation |
| Pyk2 (phospho-Tyr881) | 0.38 | 0.36–0.41 | 0.55 | 0.51–0.59 | 1.44 | Cell growth, altered |
| Raf1 (phospho-Ser259) | 1.09 | 1.01–1.16 | 1.6 | 1.5–1.71 | 1.49 | Cell growth, altered |
| Ras-GRF1 (phospho-Ser916) | 0.33 | 0.32–0.33 | 0.47 | 0.45–0.48 | 1.41 | Cell proliferation |
| RSK1/2/3/4 (phospho-Ser221/227/218/232) | 0.84 | 0.81–0.86 | 1.07 | 1.04–1.11 | 1.27 | Enzymatic activity, induced |
| STAT1 (phospho-Tyr701) | 1.12 | 1.08–1.16 | 1.57 | 1.48–1.66 | 1.41 | Cell growth, altered |
| STAT3 (phospho-Tyr705) | 0.88 | 0.86–0.9 | 1.07 | 1.05–1.09 | 1.22 | Ubiquitination |
ERK, extracellular signal-regulated kinase; MV, microvesicle; CI, confidence interval.
Table II.
Decrease in phosphorylation of ERK signaling pathway due to incubation of RSC96 cells with ACC-2 MVs.
| RSC96 | RSC96+MV | RSC96+MV/RSC96 | ||||
|---|---|---|---|---|---|---|
| Phosphorylation site | Ratio | 95% CI | Ratio | 95% CI | Ratio | Biological effects |
| BAD (phospho-Ser112) | 3.12 | 2.78–3.46 | 1.21 | 1.14–1.29 | 0.39 | Apoptosis |
| BAD (phospho-Ser155) | 1.99 | 1.85–2.13 | 1.48 | 1.3–1.65 | 0.74 | Molecular association, regulation |
| CREB (phospho-Ser142) | 1.92 | 1.84-2 | 1.36 | 1.3–1.42 | 0.71 | Activation |
| Elk1 (phospho-Ser389) | 1.81 | 1.7–1.91 | 1.38 | 1.29–1.48 | 0.76 | Intracellular localization |
| HistoneH3.1(phospho-Ser10) | 5.61 | 5.36–5.87 | 1.98 | 1.87–2.1 | 0.35 | Molecular association, regulation |
| IkB-α (phospho-Ser32/36) | 2.19 | 2–2.37 | 1.07 | 0.95–1.18 | 0.48 | Protein degradation |
| MKK6 (phospho-Ser207) | 1.71 | 1.53–1.9 | 0.59 | 0.52–0.66 | 0.35 | Positive regulation of apoptosis |
| MSK1 (phospho-Ser376) | 1.65 | 1.53–1.76 | 1.28 | 1.13–1.44 | 0.80 | Positive regulation of histone phosphorylation |
| PKA CAT (phospho-Thr197) | 0.73 | 0.71–0.76 | 0.52 | 0.51–0.53 | 0.71 | Inhibition of apoptosis |
| PLC-β (phospho-Ser1105) | 2.48 | 2.35–2.6 | 1.15 | 1.08–1.22 | 0.46 | Phospholipid catabolic process |
| Pyk2 (phospho-Tyr580) | 2.37 | 1.91–2.83 | 0.89 | 0.87–0.92 | 0.41 | Molecular association, regulation |
| Rac1/cdc42 (phospho-Ser71) | 6.35 | 5.8–6.9 | 1.03 | 0.95–1.1 | 0.16 | Cell motility involved in cell locomotion |
| Raf1 (phospho-Ser338) | 4.71 | 4.57–4.84 | 1.08 | 1.05–1.12 | 0.23 | Enzymatic activity, induced |
| Raf1 (phospho-Tyr341) | 1.23 | 1.15–1.3 | 0.71 | 0.67–0.75 | 0.58 | Enzymatic activity, induced |
| STAT3 (phospho-Ser727) | 2.39 | 2.26–2.51 | 1.62 | 1.6–1.64 | 0.68 | Activation |
ERK, extracellular signal-regulated kinase; MV, microvesicle; CI, confidence interval.
Discussion
To the best of our knowledge, the present study demonstrated for the first time that SACC cells produce MVs. With the use of electron microscopy and immunoblotting techniques, ACC-2-derived MVs were identified and characterized. Morphological analyses performed on MVs derived either from cell lines or body fluids, including urine, blood, malignant pleural effusions and amniotic fluid, as well as the present electron microscopy findings, have revealed that MVs share common characteristics, including a round shape, membranes with lipid bilayers and a diameter of 30–100 nm (23). Western blot analysis revealed the expression of MHC-I and HSP70, which are two of the typical molecular markers of MVs. The mechanism through which cells undergo MV secretion is not entirely known, although evidence has been provided that, microvesicular formation follows an ordered pattern of membrane protrusion, budding and finally detachment of spherical fragments from the cell surface (23). Furthermore, it is becoming increasingly clear that the networks of intercellular communication via MVs have the potential to affect processes as diverse as immunoregulation, cell differentiation, chemotherapy resistance, inflammation, coagulation, angiogenesis and metastasis (14,15).
The mRNA profiles present in ACC-2 cells and their MVs were characterized. Therefore, differentially expressed genes between them may elucidate the potential biological functions of ACC-2 MVs. A total of 1,355 genes with altered expression levels (>2-fold) were identified. Gene Ontology analysis revealed that a number of these mRNA transcripts associated with cell adhesion, cytoskeleton, cell defense, cell metabolism, development, cell cycle and signal transduction were present at high levels in the MVs. Identification of RNAs being present in MVs supports the hypothesis that MVs may represent a vehicle by which one cell communicates with another, delivering RNA and, in turn, modulating recipient-cell protein production (24). The results, which demonstrated that certain genes exhibited increased expression in exosomes compared with the derived cell, prompts the notion that carcinoma cells may affect other cells through selective enrichment of characteristic substances in the exosomes. This may be a novel and effective way by which cells communicate with each other. Nonetheless, the suggestion that MV cargos may be delivered to other cells provides the foundation for the proposed transfer mechanism by MVs.
Therefore, the expression of four differentially expressed genes was validated by RT-qPCR. Although the functional annotation for the four genes in oral cancer is not complete, these genes may be associated with SACC progression and metastasis according to records in Entrez Gene (25). IL-19, an important immune regulatory factor, is involved in tumor immune suppression, and may induce the mononuclear cell secretion of tumor necrosis factor-α, which may suppress the process of tumorigenesis. Therefore, with a small amount of IL-19 expression, the MVs will exhibit a hypoallergenic phenotype, thus evading an immune reaction. PDGFRA is involved in metastasis and angiogenesis in cancer; its mutation is associated with multiple types of tumor, including glioma, ovarian cancer, colorectal cancer, breast cancer and prostate cancer (26). The state of the PDGFRA gene loaded into exosomes requires additional studies for detection, but there is reason to consider that it may exert important functions subsequent to being transfected into cells through exosomes (26). HLA-DOB, which belongs to the HLA class IIβ chain paralogues, is involved in tumor antigen presentation, and the differential expression of this gene implies that exosomes may be antigen-presenting bodies that are involved in regulating immune function. HLA-DOB may alternatively suppress or improve immune function in different situations (27). In the present study, it was implied that the former is more likely; FGF12, a member of the fibroblast growth factor family, possessing mitogenic and cell-survival activities, is involved in repairment and regeneration of neural injury. FGF12 may promote the survival of the ganglia and peripheral neurons and the growth of neurites in vitro, and may promote the repair and regeneration of damaged neurons. Therefore, the increased expression of FGF12 in ACC-2 MVs may be involved in the process of perineural invasion. Additional future studies may identify important factors for the treatment of SACC (26–29). A previous study suggested that ACCs may exhibit similarities with HeLa cells (30). However, the cells used in the present study exhibited specific perineural invasion characteristics, without any resemblance to HeLa cells. Additionally, the present study demonstrated almost identical results with the cell SACC-83 (31), and ACCs have already been used previously in certain experiments to compare differential gene expression profiles associated with metastasis (32).
Finally, the DiI fluorescent probe was used to detect the interaction between the exosomes and the RSC96 cells. The results revealed that no light was observed on the surface of the cell membranes, while visable fluorescence appeared in the cells. This indicated that the exosomes were taken up by the RSC96 cells successfully and therefore, exosomes may be a good choice for loading and delivering of impartant materials and cell signals, including protein, RNA and microRNA. The phosphorylation status of direct and indirect ERK targets was then determined in ACC-2 MV-transduced RSC96 cells using a phospho-specific antibody microarray. The ERK signaling pathway is well-known to be associated with several mechanisms of tumor progression and metastasis (33). The results of the present study demonstrated that the phosphorylation of the upstream proteins and kinases of the ERK pathway were altered, and a number of these proteins were associated with cell growth, angiogenesis, cell defferentiation, inhibitation of apoptosis, development of nervous system and signal transduction, which are important for the progression, metastasis and invasion of malignant cells. Thus, ACC-2 MVs appear to affect Schwann cell lines by activation of the ERK signaling pathway. These results also provide additional in-depth understanding of the molecular targets by which SACC cell-derived MVs are involved in perineural invasion in SACC, and lay a foundation for further exploration of this novel method of tumor metastasis and invasion. A previous article has stated that ACCs may have some relationships with Hela cells. However the cells used in this study have the specific characteristic of perineural invasion without any affection with Hela cells. Furthermore, almost the same results have been observed with the SACC-83 cell line.
In conclusion, the present study demonstrated that SACC cell-derived MVs may be involved in the pathogenesis of perineural invasion in SACC, and a promising tumor biological therapeutic target. In addition, the application of exosomes as substitutes for viruses for transferring substances is worth future attention, with the complete and effective absorption of exosomes in the present study providing a basis for this research.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant nos. 81172578 and 81472532). The authors would like to thank Mrs. Xiaoyu Li from the State Key Laboratory of Oral Diseases, Sichuan University (Chengdu, China) for her skillful technical assistance.
References
- 1.Kokemueller H, Eckardt A, Brachvogel P, Hausamen JE. Adenoid cystic carcinoma of the head and neck-a 20 years experience. Int J Oral Max Surg. 2004;33:25–31. doi: 10.1054/ijom.2003.0448. [DOI] [PubMed] [Google Scholar]
- 2.Barrett AW, Speight PM. Perineural invasion in adenoid cystic carcinoma of the salivary glands: A valid prognostic indicator? Oral Oncol. 2009;45:936–940. doi: 10.1016/j.oraloncology.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Zhang P, Ma Q, Kong L, Li Y, Liu B, Lei D. EMMPRIN silencing inhibits proliferation and perineural invasion of human salivary adenoid cystic carcinoma cells in vitro and in vivo. Cancer Biol Ther. 2012;13:85–91. doi: 10.4161/cbt.13.2.18455. [DOI] [PubMed] [Google Scholar]
- 4.Atay S, Gercel-Taylor C, Kesimer M, Taylor DD. Morphologic and proteomic characterization of exosomes released by cultured extravillous trophoblast cells. Exp Cell Res. 2011;317:1192–1202. doi: 10.1016/j.yexcr.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Lee TH, D'asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer-the emerging science of cellular ‘debris’. Semin Immunopathol. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 6.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Simons M, Raposo G. Exosomes-vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Al-Nedawi K, Meehan B, Rak J. Microvesicles: Messengers and mediators of tumor progression. Cell Cycle. 2009;8:2014–2018. doi: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- 10.Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, Menna E, Saglietti L, Schuchman EH, Furlan R, Clementi E, et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009;28:1043–1054. doi: 10.1038/emboj.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Conde I, Shrimpton CN, Thiagarajan P, López JA. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood. 2005;106:1604–1611. doi: 10.1182/blood-2004-03-1095. [DOI] [PubMed] [Google Scholar]
- 12.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 13.Baran J, Baj-Krzyworzeka M, Weglarczyk K, Szatanek R, Zembala M, Barbasz J, Czupryna A, Szczepanik A, Zembala M. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol Immunother. 2010;59:841–850. doi: 10.1007/s00262-009-0808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 15.Kim CW, Lee HM, Lee TH, Kang C, Kleinman HK, Gho YS. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res. 2002;62:6312–6317. [PubMed] [Google Scholar]
- 16.Mause SF, Weber C. Microparticles: Protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010;107:1047–1057. doi: 10.1161/CIRCRESAHA.110.226456. [DOI] [PubMed] [Google Scholar]
- 17.Sanderson MP, Keller S, Alonso A, Riedle S, Dempsey PJ, Altevogt P. Generation of novel, secreted epidermal growth factor receptor (EGFR/ErbB1) isoforms via metalloprotease-dependent ectodomain shedding and exosome secretion. J Cell Biochem. 2008;103:1783–1797. doi: 10.1002/jcb.21569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimholt RM, Urdal P, Klingenberg O, Piehler AP. Rapid and reliable detection of α-globin copy number variations by quantitative real-time PCR. BMC Hematol. 2014;14:4. doi: 10.1186/2052-1839-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 20.Mallegol J, Van Niel G, Lebreton C, Lepelletier Y, Candalh C, Dugave C, Heath JK, Raposo G, Cerf-Bensussan N, Heyman M. T84-intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology. 2007;132:1866–1876. doi: 10.1053/j.gastro.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 21.Skog J, Wurdinger T, Van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas GM, Panicot-Dubois L, Lacroix R, Dignat-George F, Lombardo D, Dubois C. Cancer cell-derived microparticles bearing P-selectin glycoprotein ligand 1 accelerate thrombus formation in vivo. J Exp Med. 2009;206:1913–1927. doi: 10.1084/jem.20082297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;219:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed KA, Xiang J. Mechanisms of cellular communication through intercellular protein transfer. J Cell Mol Med. 2011;15:1458–1473. doi: 10.1111/j.1582-4934.2010.01008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao M, Ma L, Liu Y, Qu H. Pedican: An online gene resource for pediatric cancers with literature evidence. Sci Rep. 2015;5:11435. doi: 10.1038/srep11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fallas JL, Tobin HM, Lou O, Guo D, Sant'Angelo DB, Denzin LK. Ectopic expression of HLA-DO in mouse dendritic cells diminishes MHC class II antigen presentation. J Immunol. 2004;173:1549–1560. doi: 10.4049/jimmunol.173.3.1549. [DOI] [PubMed] [Google Scholar]
- 27.Hsing CH, Cheng HC, Hsu YH, Chan CH, Yeh CH, Li CF, Chang MS. Upregulated IL-19 in breast cancer promotes tumor progression and affects clinical outcome. Clin Cancer Res. 2012;18:713–725. doi: 10.1158/1078-0432.CCR-11-1532. [DOI] [PubMed] [Google Scholar]
- 28.Mei Y, Wang Z, Zhang L, Zhang Y, Li X, Liu H, Ye J, You H. Regulation of neuroblastoma differentiation by forkhead transcription factors FOXO1/3/4 through the receptor tyrosine kinase PDGFRA. Proc Nati Acad Sci USA. 2012;109:4898–4903. doi: 10.1073/pnas.1119535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama F, Yasuda T, Umeda S, Asada M, Imamura T, Meineke V, Akashi M. Fibroblast growth factor-12 (FGF12) translocation into intestinal epithelial cells is dependent on a novel cell-penetrating peptide domain: Involvement of internalization in the in vivo role of exogenous FGF12. J Biol Chem. 2011;286:25823–258344. doi: 10.1074/jbc.M110.198267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phuchareon J, Ohta Y, Woo JM, Eisele DW, Tetsu O. Genetic profiling reveals cross-contamination and misidentification of 6 adenoid cystic carcinoma cell lines: ACC2, ACC3, ACCM, ACCNS, ACCS and CAC2. PLoS One. 2009;4:e6040. doi: 10.1371/journal.pone.0006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou J, Wang F, Liu X, Song M, Yin X. Tumor-derived exosomes enhance invasion and metastasis of salivary adenoid cystic carcinoma cells. J Oral Pathol Med. 2018;47:144–151. doi: 10.1111/jop.12654. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Liu BY, Zhang X, Zhao XG, Cao G, Dong Z, Zhang SL. Identification of differentially expressed genes in salivary adenoid cystic carcinoma cells associated with metastasis. Arch Med Sci. 2016;12:881–8. doi: 10.5114/aoms.2016.60973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bratton MR, Antoon JW, Duong BN, Frigo DE, Tilghman S, Collins-Burrow BM, Elliott S, Tang Y, Melnik LI, Lai L, et al. Gαo potentiates estrogen receptor α activity via the ERK signaling pathway. J Endocrinol. 2012;214:45–54. doi: 10.1530/JOE-12-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]