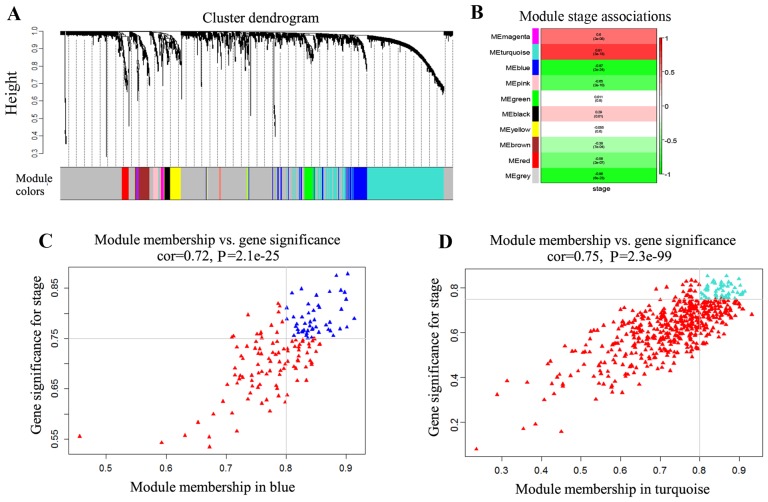
Figure 1.
Weighted gene co-expression network analysis. (A) A hierarchical clustering tree of the differential expression genes set was constructed by average linkage of hierarchical clusters. Gene set is grouped into 10 co-expression modules. (B) Heatmap of correlation between MEs and disease stages. The first value of each rectangle represents Spearman's correlation coefficient between the MEs and stages. The second value in each rectangle is the P-value. (C) Blue spots are genes with |GS|>0.75 and |MM|>0.8 in the blue module. (D) Turquoise spots are genes with |GS|>0.75 and |MM|>0.8 in the turquoise module. In (C) and (D), red spots represent the genes with |GS|<0.75 or |MM|<0.8 in the modules. ME, module eigengene; GS, gene significance; MM, module membership.