Abstract
Ovarian cancer is the most lethal gynecologic malignancy, and ovarian cancer stem cells (CSCs) serve a pivotal function in the metastasis and recurrence of ovarian cancer. Multiple previous studies have validated CD133 as a marker of ovarian CSCs. Although salinomycin is a promising therapeutic agent that has been demonstrated to kill CSCs in various types of cancer, poor aqueous solubility hampers its clinical application. The present study used salinomycin-loaded poly(lactic-co-glycolic acid)-poly(ethylene glycol) nanoparticles conjugated with CD133 antibodies (CD133-SAL-NP) to eliminate CD133+ ovarian CSCs. The results revealed that CD133-SAL-NPs were of an appropriate size (149.2 nm) and exhibited sustained drug release. CD133-SAL-NPs efficiently bound to CD133+ ovarian cancer cells, resulting in an increased cytotoxic effect in CD133+ ovarian cancer cells, compared with the untargeted SAL-NPs and salinomycin. CD133-SAL-NPs reduced the percentage of CD133+ ovarian CSCs in ovarian cells more effectively than treatment with salinomycin or SAL-NPs, suggesting that CD133-SAL-NP targeted CD133+ ovarian CSCs. In nude mice bearing ovarian cancer xenografts, CD133-SAL-NPs exerted improved therapeutic effects compared with SAL-NPs and salinomycin. Thus, CD133 was demonstrated to be a promising target for drug delivery to ovarian CSCs, and may be useful as an agent to inhibit the growth of ovarian cancer by targeting CD133+ ovarian CSCs. CD133-SAL-NPs may therefore represent a promising approach for the treatment of ovarian cancer.
Keywords: nanomedicines, ovarian cancer, cancer stem cells, salinomycin, cluster of differentiation 133
Introduction
Ovarian cancer is the most lethal gynecologic malignancy, with a majority of cases being diagnosed following metastasis of the disease (1). Due to the acquisition of chemoresistance by residual ovarian cancer cells, recurrence is frequent. Ovarian cancer stem cells (CSCs) serve a pivotal function in the recurrence and metastasis of ovarian cancer (2,3). Thus, it is necessary to eliminate therapy-resistant ovarian CSCs (4). Cluster of differentiation (CD)133 is a common CSC marker in human solid cancers, including ovarian cancer (5–8), and Baba et al (1) used a murine model to demonstrate that CD133+ ovarian cancer cells have similar characteristics to ovarian CSCs in terms of self-renewal, differentiation and tumorigenicity.
Gupta et al (9) screened a series of chemicals to discover compounds that selectively target and inhibit breast CSCs, and demonstrated that salinomycin, a polyether ionophore antibiotic, selectively inhibited CSCs and exerted potent effects against various types of CSC (9). The mechanisms underlying the anti-CSC activity of salinomycin include inhibition of the differentiation of CSCs, the Wnt/β-catenin pathway and autophagy (9,10). These results suggest that salinomycin represents a promising agent capable of targeting CSCs. To the best of our knowledge, no previous reports have investigated the therapeutic efficacy of salinomycin against ovarian CSCs. Thus, it is necessary to explore the therapeutic efficacy of salinomycin against ovarian CSCs.
There is another important issue with respect to salinomycin, which has to be resolved prior to clinical application. Owing to its poor water solubility, salinomycin must be dissolved in ethanol prior to administration (11,12). Nanoparticles have emerged as a promising approach to improve the solubility of salinomycin (5,6). Several salinomycin-loaded nanoparticles have been developed for its delivery to CSCs, and these have achieved an improved therapeutic effect over free salinomycin (13,14). Poly(lactic-co-glycolic acid) (PLGA) nanoparticles are widely used due to their safety record in humans (15,16). PLGA nanoparticles are frequently modified with poly(ethylene glycol) (PEG) to increase their in vivo circulation. The PEGylation of nanoparticles significantly increases their passive targeting of tumors (15).
Extensive interest has been generated in antibody-conjugated nanoparticles, as these are widely used as targeted drug delivery systems (17). Several antibodies, including anti-human epidermal growth factor receptor 2 (HER2) or anti-epidermal growth factor receptor (EGFR) antibodies, have been used to promote the delivery of small interfering RNA nanoparticles to EGFR-overexpressing or HER2-overexpressing cancers (18,19). As cluster of differentiation (CD)133 is considered to be a marker for ovarian CSCs, our group hypothesized that the CD133 antibody may be able to promote the salinomycin delivery of nanoparticles to CD133-overexpressing ovarian CSCs.
In the present study, the CSC-like properties of purified CD133+ cells from the OVCAR-3 and PA-1 ovarian cancer cell lines were demonstrated. Subsequently, salinomycin-loaded PLGA nanoparticles conjugated with CD133 antibodies (CD133-SAL-NP) were developed to target CD133+ ovarian CSCs.
Materials and methods
Materials
Poly(lactide-co-glycolide)-b-poly(ethylene glycol)-COOH endcap (PLGA-PEG-COOH; ~17,000 Da, 3,400 Da) was purchased from Akina, Inc. (West Lafayette, IN, USA). Phycoerythrin (PE)-conjugated CD133 antibodies (cat. no. 130080801) and the CD133 MicroBead kit were provided by Miltenyi Biotec, Inc. (Auburn, CA, USA). CD133 antibodies were purchased from R&D Systems, Inc. (cat. no. MAB11331; Minneapolis, MN, USA). Epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), B-27, insulin-transferrin-selenium (ITS), and TRIzol reagent were provided by Thermo Fisher Scientific, Inc. (Waltham, MA, USA). N-hydroxysuccinimide (NHS), 1-ethyl-3-3-dimethylaminopropyl carbodiimide (EDC), salinomycin, coumarin 6 and all analytical grade organic reagents were purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Cell Counting Kit-8 was purchased from Dojindo Molecular Technologies, Inc. (Kumamoto, Japan), and the Reverse Transcription System kit was provided by Promega Corporation (Madison, WI, USA).
Cells and culture
The OVCAR-3 and PA-1 ovarian cancer cell lines (American Type Culture Collection, Manassas, VA, USA) were maintained at 37°C in RPMI-1640 medium (Thermo Fisher Scientific, Inc.) supplemented with L-glutamine and 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Inc.) in an atmosphere of 5% CO2/95% air.
Analysis of CD133 expression
Flow cytometry was used to evaluate CD133 expression in ovarian cancer cells. The cells (1×105) were trypsinized with 0.05% Trypsin (Thermo Fisher Scientific, Inc.) washed with PBS (pH 7.4; Thermo Fisher Scientific, Inc.), and centrifuged at 1,000 × g for 5 min at 4°C. Collected cells were then incubated with 1 µg/ml PE-CD133 antibodies (1:100 dilution, cat. no. 130080801) diluted in 1% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) for 30 min at 4°C. CD133 expression was analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA). Expression was analysed using FlowJo (version 10; FlowJo LLC, Ashland, OR, USA).
Isolation of CD133+ cells from ovarian cancer cell lines
CD133+ cells were isolated from cancer cells using the protocol provided by the CD133 MicroBead kit (Miltenyi Biotec, Inc.). The cell suspension in PBS (pH 7.4) was incubated with CD133 microbeads for 20 min at 4°C. Following two washes with PBE [PBS with 0.5% bovine serum albumin (Miltenyi Biotec, Inc.) and 5 mM EDTA], the unbound microbeads were removed from the cells, and 0.5 ml PBE was added. Following mixing, the cells were separated using a magnetic separation column (Miltenyi Biotec, Inc.) according to the manufacturer's protocol, and the CD133+ cells retained by the column were eluted with 0.2 ml PBE and collected. The purity of the CD133+ cells was determined using flow cytometry as aforementioned.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted using TRIzol reagent according to the manufacturer's protocol. First-strand cDNA was reverse transcribed using the Reverse Transcription System kit according to the manufacturer's protocol. The temperature protocol of reverse transcription was as follows: 70°C for 10 min, 42°C for 15 min, 95°C for 5 min and 4°C for 5 min. SYBR™-Green PCR Master Mix (Thermo Fisher Scientific, Inc.) and a Roche Light Cycler (Roche Diagnostics GmbH, Mannheim, Germany) was used to perform PCR. The sequence of the primers were as follows: β-actin forward, 5′-CGTGGACATCCGTAAAGACC-3′ and reverse, 5′-ACATCTGCTGGAAGGTGGAC-3′; CD133 forward, 5′-TCAATTTTGGATTCATATGCCTT-3′ and reverse, 5′-ACTCCCATAAAGCTGGACCC-3′; NG2 forward, 5′-TTGGCTTTGACCCTGACTATG-3′ and reverse, 5′-CTGCAGGTCTATGTCGGTCA-3′; OCT4 forward, 5′-GCGAACCATCTCTGTGGTCT-3′ and reverse, 5′-CCCCCTGTCCCCCATTCCTA-3′; SMO forward, 5′-GGCATGTATACGGCACACAG-3′ and reverse, 5′-CACCTCCACACTGCTGGC-3′; NANOG forward, 5′-GATTTGTGGGCCTGAAGAAA-3′ and reverse, 5′-TTGGGACTGGTGGAAGAATC-3′; and C-MYC forward, 5′-GCTGCTTAGACGCTGGATTT-3′ and reverse, 5′-CTCCTCCTCGTCGCAGTAGA-3′. After 2 min of denaturation at 95°C, 40 PCR cycles were performed with 3 sec denaturation at 95°C, 10 sec annealing at 55°C and 25 sec extension at 72°C. mRNA expression was quantified using the 2ΔΔCq method (20).
Development of salinomycin-loaded PLGA nanoparticles
The emulsion/solvent evaporation approach was used to create SAL-NPs, as previously described (15). PLGA-PEG-COOH (30 mg) and salinomycin (5 mg) were dissolved in dichloromethane (2 ml) to form the drug solution, which was added into an aqueous sodium cholate solution (1%; 4 ml). Following sonication using a probe sonicator (45 sec; 150 W), the resultant emulsion was added into sodium cholate solution (0.5%; 20 ml). Following evaporation, dichloromethane was discarded from the emulsion. The unencapsulated salinomycin was removed by ultrafiltration (nominal molecular weight limit, 100,000) in deionized water. Blank nanoparticles were fabricated as aforementioned, except without the addition of salinomycin. For the fabrication of coumarin-6-loaded nanoparticles (C6-NPs), 0.1% (w/w) coumarin 6 was added as aforementioned.
The development of CD133-SAL-NP was completed by the conjugation of the CD133 antibody to SAL-NPs (16). A solution of SAL-NPs (2.5 mg/ml) was activated with 50 mM NHS and 100 mM EDC for 60 min at 25°C, and the activated SAL-NPs was washed with PBS twice, incubated with CD133 antibodies (1 ml; 0.05 mg/ml; 1:10) for 6 h at 25°C to produce CD133-SAL-NP, which were washed and stored at 4°C until use.
The following abbreviations are used: SAL-NPs, salinomycin-loaded PLGA nanoparticles; CD133-SAL-NPs, salinomycin-loaded PLGA nanoparticles with CD133 antibodies; C6-NPs, coumarin 6-loaded PLGA nanoparticles; CD133-C6-NPs, coumarin 6-loaded PLGA nanoparticles with CD133 antibodies; and CD133-NPs, blank PLGA nanoparticles with CD133 antibodies.
Characteristics of nanoparticles
A dynamic light-scattering Zetasizer Nano-ZS (Malvern Instruments Ltd., Malvern, UK) was used to analyze the size and ζ potential of the nanoparticles suspended in deionized water. A total of 10 µl of the nanoparticles were mixed with 100 µl 2% phosphotungstic acid, and incubated at room temperature for 5 min. Following this, 10 µl of the nanoparticle sample was put onto a carbon-coated 400-mesh copper grid of 25 µm and allowed the dry. The morphology of nanoparticles was observed by H-600 transmission electron microscopy (TEM; Hitachi, Ltd., Tokyo, Japan).
The analysis of in vitro drug release of nanoparticles was performed as follows. Nanoparticles (1 mg/ml) were placed in a centrifuge tube, which was placed on an orbital shaker (80 × g, 37°C for 300 h). The release medium was PBS (pH 7.4) or PBS supplemented with 10% FBS. The tubes were centrifuged (20,000 g × 20 min) at 25°C after different lengths of time (1, 2, 4, 8, 24, 48, 72, 144, 216 and 288 h). Fresh medium (PBS only or PBS supplemented with 10% FBS) was added to the pellet, and the supernatant was analyzed by high-performance liquid chromatography (HPLC) as described in the following section. Briefly, the supernatant was evaporated, and 5 ml MeOH was added to form a clear sample suitable for HPLC analysis.
Salinomycin loading of nanoparticles
Nanoparticles (10 mg) were ruptured in dichloromethane for 30 min at 25°C, which was then evaporated, and 5 ml MeOH was added to form a clear sample suitable for HPLC analysis. The HPLC system (L-2000; Hitachi, Ltd.) comprised a reverse phase C-18 column (250×4.6 mm; 5 µm) with a mobile phase of phosphoric acid/tetrahydrofuran/deionized water/acetonitrile (0.01/5/10/85, v/v), a flow rate of 1.5 ml/min, and a detection wavelength of 210 nm. The column temperature was set to 35°C. Salinomycin encapsulation efficacy = the mass of salinomycin loaded/the mass of total salinomycin added ×100%. Salinomycin loading = the mass of salinomycin loaded/the mass of nanoparticles ×100%. A standard curve of coumarin 6 was used to determine the drug loading of coumarin 6.
In vitro uptake in ovarian cancer cells
CD133+ or CD133− ovarian cancer cells were inoculated (2×105 cells/well) in 12-well tissue culture plates overnight at 37°C. The cells were then treated with free coumarin 6 or coumarin 6-loaded nanoparticles (25 ng/ml coumarin 6) at 37°C. After 2 h treatment, the cells were washed with PBS supplemented with 0.1% Tween-20 and analyzed by flow cytometry (BD Biosciences). Data was analyzed using FlowJo (version 10; FlowJo LLC, Ashland, OR, USA).
CCK-8 assay
Cells were inoculated (5×103 cells/well) in 96-well plates overnight at 37°C, and different concentrations of nanoparticles or free salinomycin (0.01, 0.04, 0.12, 0.37, 1.11, 3.33, 10.00, 30.00, 90.00 and 270.00 µg/ml) were added and incubated with the cells for 48 h. Finally, cell viability was assessed using CCK-8 according to the manufacturer's protocol.
Effect of nanoparticles on CSC percentage
The effect of nanoparticle treatment on the CSC percentage of ovarian cancer cells was examined by determining tumor sphere formation ability and the percentage of CD133+ cells. Ovarian cancer cells (5×104 cells/well) were inoculated in RMPI-1640 medium with L-glutamine and 10% FBS in 12-well plates and left overnight. Following this, nanoparticles (60 µg/ml) or free salinomycin (5 µg/ml) were added to the cells and incubated for 24 h at 37°C. The drugs were then removed and discarded, the cells were washed with PBS twice, and fresh medium was added for a further 72 h. Thereafter, the collected cells were cultured in ultra-low adherent 6-well dishes (2,000 cells/well; Corning, Tewksbury, MA, USA) to obtain tumorspheres. The cells were suspended in stem cell culture medium [1X ITS, 1X B27 (Invitrogen; Thermo Fisher Scientific, Inc.), 20 ng/ml bFGF and 20 ng/l EGF]. After 7 days, the number of tumor spheres was counted under a light microscope, and three fields of view were assessed in each group. Alternatively, the cells were detached by incubation with trypsin, and the percentage of CD133+ cells was measured.
Animal studies
The experimental protocols were approved by the Institutional Animal Care and Use Committee of Wuhan University (Wuhan, China).
In vivo tumorigenicity of ovarian cancer cells
CD133+ or CD133− ovarian cancer cells were mixed with BD Matrigel (growth factor-reduced Matrigel; BD Biosciences) and subcutaneously implanted into the right flank of 6-week-old female BALB/c nude mice (weight, ~20 g, 6 mice per group). Animals were housed in separate cages (3–4 animals per cage) maintained under a controlled atmosphere (humidity of 50±7% and temperature of 21±1°C) and with a 12:12-h light/dark cycle. The mice were allowed free access to food and water. Mice were euthanized if the tumor size exceeded 1,500 mm3. Tumor formation was monitored over 20 weeks.
In vivo antitumor assay
PA-1 cells (2×106) were subcutaneously inoculated into the right flank of the 6-week-old female BALB/c nude mice (weight, ~20 g, 8 mice per group) on day 0. The mice were maintained as aforementioned. On day 10, the average tumor volume reached ~50 mm3. Starting from day 10, nanoparticles were intravenously administered to the mice (5 mg salinomycin/kg) via the tail vein. Free salinomycin was dissolved in ethanol and administered by intraperitoneal injection. The drugs were administered once every 2 days, a total of 9 times. The tumor volume was calculated as follows: Width2 × length)/2. On day 30, the effect of the drugs on the percentage of CSCs was evaluated by the following method. After the mice were euthanized, the tumors were excised, washed with PBS, chopped, digested with a collagenase I solution, and filtered using a cell strainer (40-µm). The strained cells were cultured overnight, and the percentage of CSCs was evaluated as aforementioned.
Statistical analysis
Data were analyzed using SPSS (version 13; SPSS, Inc., Chicago, IL, USA). Independent samples Student's t-tests were used to compare the means of two groups, and one-way analysis of variance followed by Student-Newman-Keuls post hoc tests or Dunnett's post hoc tests was used to compare the means of three or more groups. P<0.05 was considered to indicate a statistically significant difference.
Results
CD133+ ovarian cancer cells possess the properties of ovarian CSCs
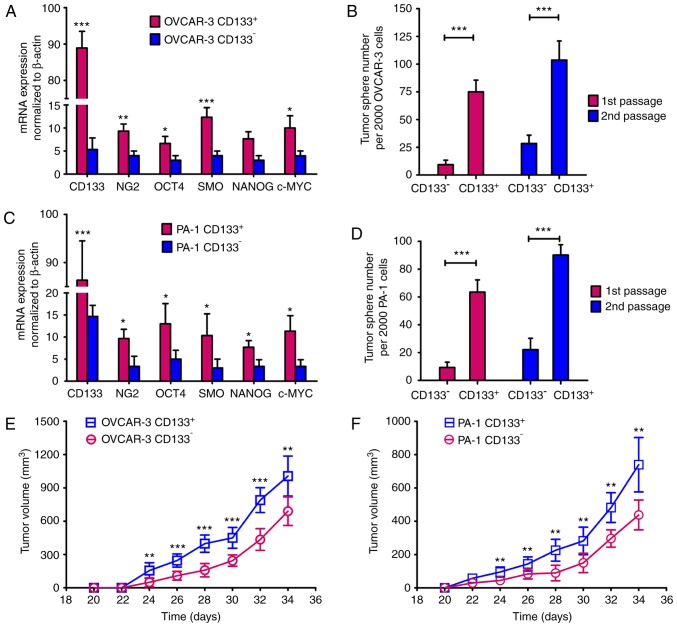
The percentage of CD133+ cells in the ovarian cancer cell lines was ~20%. Following CD133-based cell sorting, a population of >98% CD133+ cells was obtained. A series of experiments were then performed to demonstrate whether CD133+ ovarian cancer cells possessed the properties of ovarian CSCs (Fig. 1; Table I). First, the mRNA expression levels of CSC-associated genes were measured (Fig. 1A and C). CD133, ring finger protein 5, smoothened, frizzled class receptor, POU class 5 homeobox 1, NANOG and c-MYC were revealed to be significantly increased in CD133+ cells compared with CD133− cells (P<0.05). The tumor sphere formation ability represents the self-renewal capability of CSCs (11,12). As presented in Fig. 1B and D, the number of tumor spheres formed by CD133+ cells was increased compared with CD133− cells (P<0.001). As demonstrated by the tumor growth curves (Fig. 1E and F), CD133+ cells formed larger tumors than CD133− cells (P<0.05). Furthermore, when the cell count was ≥1×104, the tumor incidence rate in the CD133+OVCAR-3 group was 100% (Table I). In contrast, tumors were induced in only 25% of CD133−OVCAR-3 group, even at a cell number of 1×105, which suggested that CD133+OVCAR-3 cells have increased tumorigenic potential compared with CD133−OVCAR-3 cells (Table I). Similar results were observed with CD133+ and CD133−PA-1 cells: When the cell count was ≥1×104, the tumor incidence rate in the CD133+PA-1 group was 100%; however, even with 1×105 cells, tumors were induced in only 37.5% of the CD133−PA-1 group, indicating that CD133+PA-1 cells had increased tumorigenic potential compared with CD133−PA-1 cells (Table I). Collectively, these results demonstrated that CD133+ ovarian cancer cells exhibit potent self-renewal abilities and tumorigenicity, corroborating the suggestion that CD133+ ovarian cancer cells have similar properties to ovarian CSCs.
Figure 1.
CD133+ ovarian cancer cells have ovarian cancer stem cell properties. (A) The mRNA expression levels in OVCAR-3 ovarian cancer cells were analyzed by RT-qPCR (B) CD133+ OVCAR-3 ovarian cancer cells generated more tumor spheres than CD133− OVCAR-3 ovarian cancer cells. (C) The mRNA expression levels in PA-1 ovarian cancer cells were analyzed by RT-qPCR (D) CD133+ PA-1 ovarian cancer cells generated more tumor spheres than CD133− PA-1 ovarian cancer cells. Data are expressed as the mean ± SD (n=3). Tumor growth curves for (E) OVCAR-3 and (F) PA-1 ovarian cancer cells. CD133+ or CD133− ovarian cancer cells (5×105) were inoculated in BALB/c nude mice. The data are expressed as the mean ± SD (n=6). All data were analyzed using independent samples Student's t-tests. *P<0.05, **P<0.01 and ***P<0.001 vs. CD133−. CD, cluster of differentiation; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SD, standard deviation.
Table I.
In vivo tumorigenic potential of CD133+ or CD133-OVCAR-3 and PA-1 cells.a
| Number of cells | ||||||
|---|---|---|---|---|---|---|
| Cell type | 1×106 | 1×105 | 5×104 | 1×104 | 5×103 | 2×103 |
| CD133−OVCAR-3 | 6/8 | 2/8 | 1/8 | 1/8 | 0/8 | 0/8 |
| CD133+OVCAR-3 | 8/8 | 8/8 | 8/8 | 8/8 | 7/8 | 6/8 |
| CD133−PA-1 | 7/8 | 3/8 | 2/8 | 1/8 | 0/8 | 0/8 |
| CD133+PA-1 | 8/8 | 8/8 | 8/8 | 8/8 | 6/8 | 5/8 |
CD133+ or CD133− ovarian cancer cells were collected, mixed with growth factor-reduced Matrigel and implanted subcutaneously into BALB/c nude mice. Tumor formation was examined over a period of 20 weeks. CD, cluster of differentiation.
Nanoparticle characteristics
CD133-SAL-NPs were 149.2 nm in size, and were slightly larger than the SAL-NPs (Table II and Fig. 2A). As CD133-SAL-NPs were larger than the SAL-NPs, this indicated that antibody modification increased the size of the nanoparticles. The nanoparticles possessed a small polydispersity of 0.2, which was indicative of the narrow size distribution of the nanoparticles. The prepared nanoparticles had a relatively high, negative ζ potential (~-20 mV), which suggested that they may have high stability in circulation. The nanoparticle drug loading was ~9% and the encapsulation efficiency was ~70%, suggesting that the emulsion/solvent evaporation method was effective for the encapsulation of hydrophobic salinomycin in polymeric nanoparticles. Collectively, the prepared nanoparticles possessed an appropriate size, ζ potential, and drug-loading capacity required for useful drug delivery systems.
Table II.
Characterization of nanoparticles.
| Nanoparticle | Size (nm) | ζ potential (mv) | PDI | Drug loading (%) | EE (%) |
|---|---|---|---|---|---|
| SAL-NP | 139.9±22.9 | −19.6±6.8 | 0.16±0.07 | 9.9±2.5 | 68.3±7.8 |
| CD133-SAL-NP | 149.2±28.7 | −22.8±7.9 | 0.18±0.09 | 8.5±1.9 | 63.2±7.5 |
Data are expressed as the mean ± standard deviation (n=3). PDI, polydispersity; EE, encapsulation efficacy; SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticles; CD133-SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticles conjugated to cluster of differentiation 133 antibodies.
Figure 2.
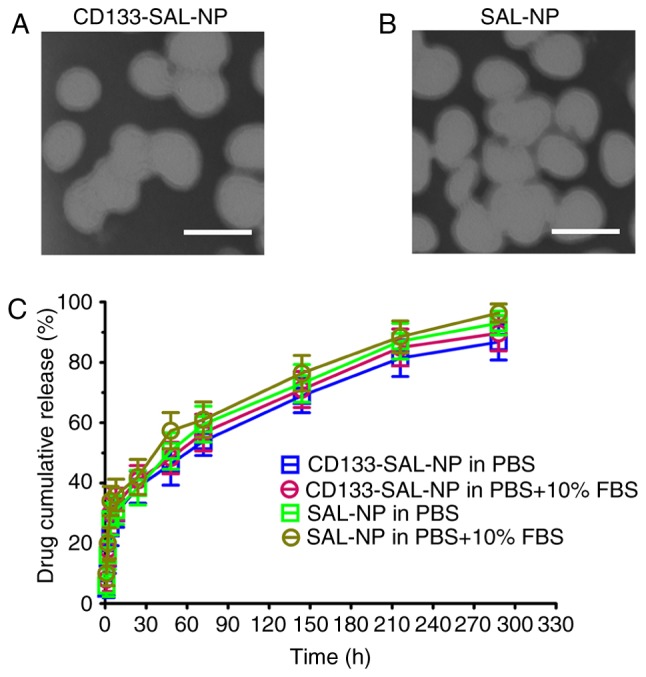
Transmission electron microscopy. Representative images of (A) CD133-SAL-NPs and (B) SAL-NPs. Scale bars, 100 nm. (C). Release of salinomycin from the nanoparticles in PBS and PBS + 10% FBS. The data are expressed as the mean ± standard deviation (n=3). CD133-SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies; SAL-NP, salinomycin-loaded poly (lactic-co-glycolic acid) nanoparticle; FBS, fetal bovine serum.
The TEM images revealed that the nanoparticles were spherical, with a relatively monodisperse size of ~100 nm (Fig. 2A and B). The size of the nanoparticles calculated from TEM observations was smaller than that calculated using measurements from dynamic light scattering; dynamic light scattering reflects the hydrodynamic size of nanoparticles, but TEM reflects the size of dried nanoparticles.
The in vitro release of nanoparticles was also measured (Fig. 2B). The nanoparticles had similar release patterns in the two release media, with fast salinomycin release observed in both nanoparticles (~45%) in the initial 24 h. A cumulative release of ~60% occurred within the following 48 h. Total release of the nanoparticles (~80%) occurred over 12 days. Thus, the two types of nanoparticle showed sustained drug release over a period of 12 days.
In vitro cellular uptake
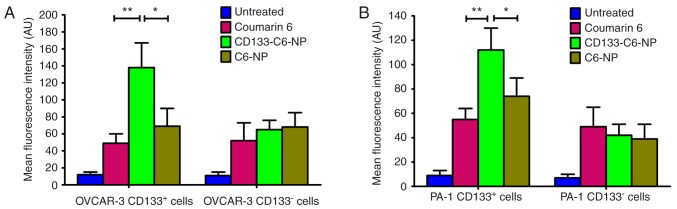
The in vitro cellular uptake of nanoparticles was evaluated by flow cytometry using coumarin 6 as a fluorescent marker (Fig. 3A). In CD133+OVCAR-3 cells, the mean fluorescence intensity (MFI) in the CD133-C6-NP-treated group was significantly increased compared with the C6-NP-treated and coumarin 6-treated groups (P<0.05). Nevertheless, no significant difference in MFI in CD133−OVCAR-3 cells was observed among the groups treated with coumarin 6, C6-NPs, and CD133-C6-NPs. Similar results were observed in CD133+ and CD133−PA-1 cells (Fig. 3B). In CD133+PA-1 cells, the MFI in the CD133-C6-NP-treated group was significantly increased (P<0.05 vs. C6-NP and coumarin-6), whereas the MFI in the CD133−PA-1 cells treated with coumarin 6, C6-NPs, and CD133-C6-NPs was similar. Thus, CD133-C6-NPs had significantly increased targeting efficacy towards CD133+ ovarian cancer cells compared with other controls.
Figure 3.
In vitro cellular uptake of nanoparticles. The CD133-C6-NP-treated group was compared with other groups by one-way analysis of variance with Dunnett's post hoc tests. (A) OVCAR-3 cells, (B) PA-1 cells. The data are expressed as the mean ± standard deviation (n=3). *P<0.05 and **P<0.01, with comparisons indicated by lines. CD133-C6-NP, coumarin 6-loaded poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies; C6-NP, coumarin 6-loaded poly(lactic-co-glycolic acid) nanoparticle; CD, cluster of differentiation.
Cell viability assays
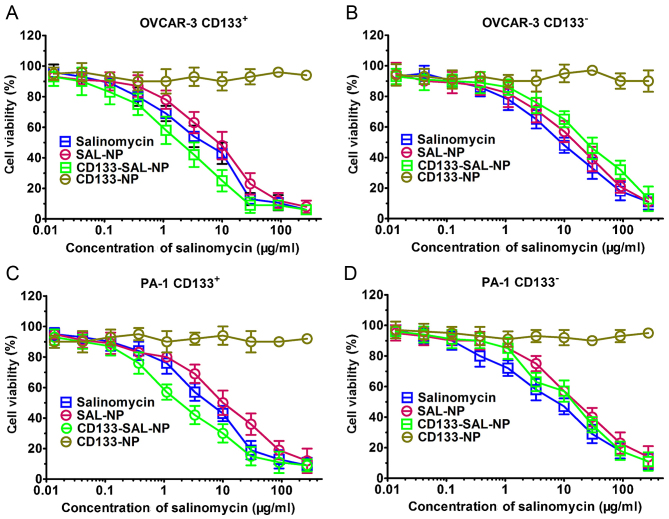
The effect of salinomycin and nanoparticles on cell viability was examined in ovarian cancer cells. As expected, CD133-NPs did not exert potent cytotoxic effects, which was reflected by >90% cell viability in the presence of CD133-NPs (Fig. 4). However, CD133-SAL-NP, SAL-NPs, and salinomycin exerted potent cytotoxicity. The half maximal inhibitory concentration (IC50) values of the drugs were calculated to quantitatively define the in vitro cytotoxicity (Table III). The IC50 value of CD133-SAL-NP (1.98 µg/ml) in CD133+OVCAR-3 cells was significantly decreased compared with that in SAL-NPs (12.34 µg/ml; P<0.001) and salinomycin (6.53 µg/ml; P<0.01). In contrast, the IC50 value of CD133-SAL-NP (19.35 µg/ml) in CD133−OVCAR-3 cells did not significantly differ from that of SAL-NPs (18.12 µg/ml) or salinomycin (13.82 µg/ml; P>0.05). CD133-SAL-NPs were 6.23- or 3.30-fold more effective compared with SAL-NPs or salinomycin in CD133+OVCAR-3 cells, respectively. Similar results were observed in CD133+PA-1 and CD133−PA-1 cells. In CD133+PA-1 cells, CD133-SAL-NPs were 3.34- or 2.06-fold more effective compared with SAL-NPs or salinomycin, respectively. Thus, the increased cytotoxicity of CD133-SAL-NPs in CD133+ ovarian cancer cells resulted from increased expression of CD133 in CD133+ ovarian cancer cells.
Figure 4.
Cell viability assays. Cell viability was assessed in (A) CD133+OVCAR-3 ovarian cancer cells, (B) CD133−OVCAR-3 ovarian cancer cells, (C) CD133+PA-1 ovarian cancer cells and (D) CD133−PA-1 ovarian cancer cells using Cell Counting Kit-8. The data are expressed as the mean ± standard deviation (n=3). CD, cluster of differentiation; SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticle; CD133-SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies; CD133-NP, poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies.
Table III.
Results of cell viability assays following nanoparticle or free drug treatment for 48 h.
| IC50 (µg/ml) | ||||
|---|---|---|---|---|
| OVCAR-3 | PA-1 | |||
| Treatment | CD133+ | CD133− | CD133+ | CD133− |
| Salinomycin | 6.53±2.71 | 13.82±6.56 | 9.12±3.45 | 11.54±6.94 |
| SAL-NP | 12.34±5.73 | 18.12±5.93 | 14.79±5.67 | 18.05±6.04 |
| CD133-SAL-NP | 1.98±1.59 | 19.35±6.44 | 4.42±2.11 | 19.58±3.43 |
| CD133-NP | >250.0 | >250.0 | >250.0 | >250.0 |
Data are expressed as the mean ± standard deviation (n=3). IC50, half maximal inhibitory concentration; CD, cluster of differentiation; SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticles; CD133-SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticles conjugated with CD133 antibodies; CD133-NP, poly(lactic-co-glycolic acid) nanoparticles conjugated with CD133 antibodies.
Influence of nanoparticles on the CSC percentage in ovarian cancer cells
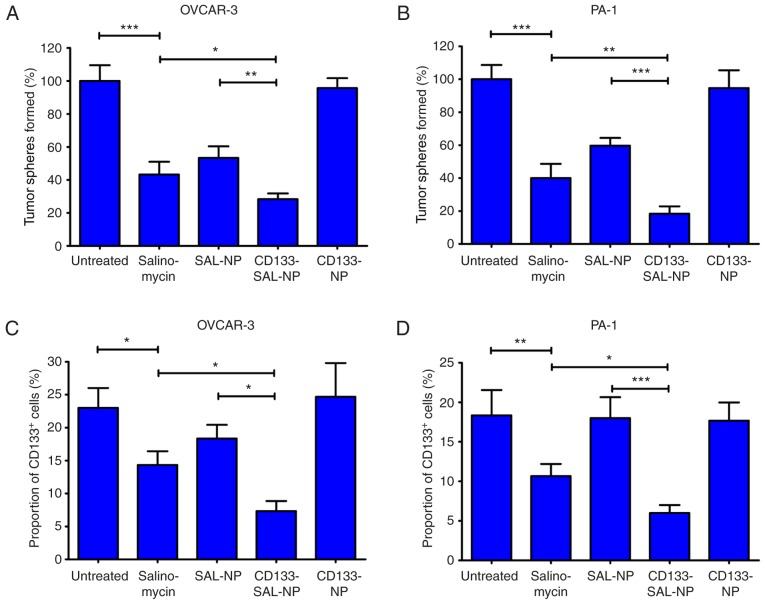
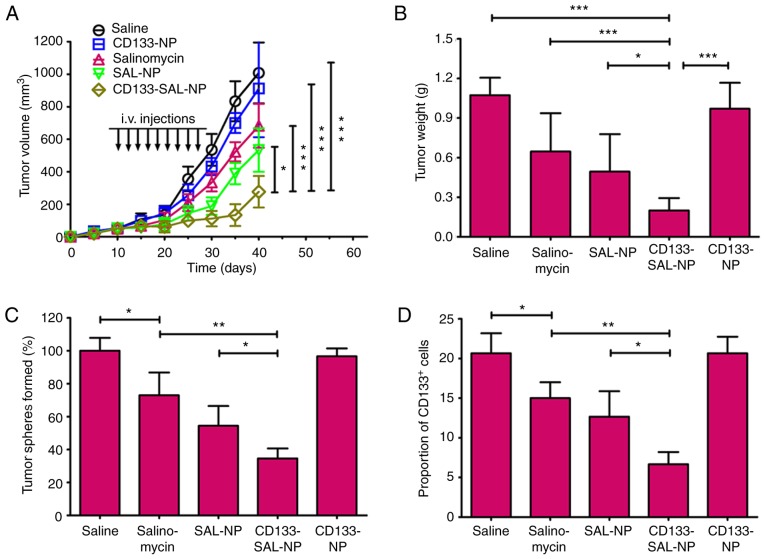
Tumor sphere formation and the percentage of CD133+ cells were measured to determine the influence of nanoparticles on the CSC percentage in ovarian cancer cells (Fig. 5). Capacity for tumor sphere formation is considered to be associated with CSC percentage (21). In OVCAR-3 cells, CD133-SAL-NPs resulted in a decreased number of tumor spheres than SAL-NPs and salinomycin (P<0.01; Fig. 5A), and a four-fold decrease compared with the untreated control. Similar results were observed concerning the percentage of CD133+ cells in OVCAR-3 cells. CD133-SAL-NPs significantly reduced the percentage of CD133+ cells in OVCAR-3 cells compared with SAL-NPs and salinomycin (P<0.05; Fig. 5C).
Figure 5.
Effect of the nanoparticles or free drugs on the cancer stem cell percentage in ovarian cancer cells. Tumor sphere formation assays were performed in (A) OVCAR-3 and (B) PA-1 cells, following the indicated treatment conditions. The tumor sphere formation rate of the untreated group was used as a control, in which the rate was defined as 100%. The percentage of CD133+ cells was analyzed in (C) OVCAR-3 and (D) PA-1 cells following the indicated treatment conditions. The groups were compared using one-way analysis of variance with Student-Newman-Keuls post hoc tests. The data are expressed as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 and ***P<0.001, with comparisons indicated by lines. CD, cluster of differentiation; SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticle; CD133-SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies; CD133-NP, poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies.
Similar results were observed in PA-1 cells. CD133-SAL-NP treatment led to a five-fold decrease in the number of PA-1 tumor spheres (Fig. 5B), which was significantly lower than that observed following treatment with SAL-NPs and salinomycin (P<0.01). Furthermore, CD133-SAL-NPs reduced the percentage of CD133+ cells in PA-1 cells significantly compared with salinomycin and SAL-NPs (P<0.05; Fig. 5D). Thus, the therapeutic efficacy of CD133-SAL-NPs in the treatment of CD133+ ovarian cancer cells was superior compared with SAL-NP and salinomycin treatment.
In vivo antitumor efficacy
The PA-1 tumors progressed rapidly in the mice in the saline- and CD133-NP-treated groups, which showed that the CD133-NPs had no antitumor effects. In contrast, salinomycin, SAL-NPs and CD133-SAL-NPs exerted antitumor effects. The fold increase of the tumor volume at the endpoint compared with the initial tumor volume was used as an index of the therapeutic efficacy of the drugs. Treatment with salinomycin and SAL-NPs resulted in 13.7- and 10.7-fold increases, respectively, whereas CD133-SAL-NPs exerted the most optimal therapeutic efficacy, with a 5.5-fold increase recorded. At the endpoint, the tumor volume of the CD133-SAL-NP-treated group was the smallest among all the groups (P<0.05; Fig. 6A) and the weight of the tumors in the CD133-SAL-NP-treated group was decreased compared with all the other groups (P<0.05; Fig. 6B). Furthermore, the tumor weight of the SAL-NP-treated group was decreased compared with the CD133-NP-treated and saline-treated groups (P<0.05).
Figure 6.
In vivo antitumor activity. (A) Tumor growth curves. (B) Excised tumors were weighed at the endpoint. Tumor volume and weight between groups were analyzed using one-way ANOVA with the Dunnett's post hoc tests. The data are expressed as the mean ± SD (n=8). The influence of the drugs on the in vivo cancer stem cell percentage was evaluated by examining (C) tumor sphere formation and (D) the percentage of CD133+ cells in the cells isolated from the excised tumors on day 30. The groups were compared using one-way ANOVA with Student-Newman-Keuls post hoc tests. The data are expressed as the mean ± SD (n=5). *P<0.05, **P<0.01 and ***P<0.001, with comparisons indicated by lines. CD, cluster of differentiation; SD, standard deviation; ANOVA, analysis of variance; SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticle; CD133-SAL-NP, salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies; CD133-NP, poly(lactic-co-glycolic acid) nanoparticle conjugated with cluster of differentiation 133 antibodies.
The population of ovarian CSCs in the excised in vivo tumors was also evaluated (Fig. 6C and D). CD133-NP did not influence the tumor sphere formation of PA-1 cells. Notably, CD133-SAL-NPs were the most active against ovarian CSCs, as reflected by the 2.5-fold decrease in the PA-1 tumor sphere number relative to the saline control (Fig. 6D). CD133-SAL-NP treatment led to significantly fewer tumor spheres than SAL-NP or salinomycin treatment (P<0.05). Salinomycin treatment led to a significantly decreased proportion of CD133+ ovarian cancer cells compared with the saline control (P<0.05). Notably, CD133-SAL-NP treatment resulted in a significantly decreased percentage of CD133+ ovarian cancer cells compared with that induced salinomycin and SAL-NP treatment (P<0.05; Fig. 6D). Taken together, these results indicate that CD133-SAL-NPs represent a promising treatment for the in vivo reduction of the ovarian CSC population.
Discussion
Improving the treatment of ovarian cancer is an urgent task in terms of public health. One of the critical reasons for reduced survival and treatment failure in ovarian cancer is the presence of ovarian CSCs. Therefore, the development of treatments specifically targeting ovarian CSCs is critical to improve ovarian cancer therapies. In the present study, CD133-SAL-NPs were demonstrated to effectively reduce the number of CD133+ ovarian CSCs.
In 2009, salinomycin was revealed to exert potent anti-CSC activity in breast cancer (8) and has since been demonstrated to be able to target and kill various types of CSC, suggesting that salinomycin represents a broad-spectrum reagent against CSCs. For example, salinomycin was evaluated in a clinical trial of a small cohort of patients with breast cancer (9). The results revealed that salinomycin achieved partial regression of tumor metastasis without severe acute side effects. Thus, salinomycin represents a highly promising drug targeting CSCs. Although salinomycin has demonstrated potent anti-CSC activity in various types of cancer, to the best of our knowledge the activity of salinomycin against ovarian CSCs has not been demonstrated in previous studies. In the present study, salinomycin was revealed to exert significantly increased cytotoxic effects in CD133+ ovarian cancer cells compared with CD133− ovarian cancer cells. Furthermore, salinomycin significantly reduced the CSC percentage in ovarian cancer cells. To the best of our knowledge, the present study is the first to report of the potent activity of salinomycin against ovarian CSCs.
Although salinomycin has demonstrated potent activity against ovarian CSCs, it has poor water solubility and low bioavailability. The hydrophobic drug loading of nanoparticles represents a practical way to increase their therapeutic efficacy (22,23). The present study used nanoparticles made from PLGA, which is a Food and Drug Administration-approved biodegradable polymer, to increase the solubility of salinomycin and improve the targeting of salinomycin towards ovarian cancer cells. Furthermore, the addition of the CD133 antibody was crucial in the targeting of CD133-SAL-NPs to CD133+ ovarian cancer cells. Flow cytometry revealed that CD133-C6-NPs were capable of efficiently binding to CD133+ ovarian cancer cells. Following binding to cells, CD133-SAL-NPs resulted in an increased cytotoxic effect in CD133+ ovarian cancer cells compared with untargeted SAL-NPs and salinomycin. In comparison with SAL-NPs and salinomycin, CD133-SAL-NP treatment resulted in a more effective decrease in the percentage of CD133+ ovarian CSCs, which indicated that CD133-SAL-NP selectively targeted CD133+ ovarian CSCs. Collectively, CD133-SAL-NPs were able to significantly increase the efficacy of salinomycin against CD133+ ovarian cancer cells.
CSCs possess a phenotype that distinguishes them from non-CSCs (24). This CSC phenotype may be used as a basis for the specific targeting of CSCs (25). Single-walled carbon nanotubes with CD133 monoclonal antibodies (anti-CD133-SWNTs), developed by Wang et al (26), have been demonstrated to block the tumorigenicity of glioblastoma-CD133+ cells. However, single-walled carbon nanotubes may present risks to human health (27). In contrast, the PLGA nanoparticles prepared in the present study are more biocompatible (22). However, the in vivo anti-CSC activity of anti-CD133-SWNTs was not investigated in this previous study (27). As CSCs account for a small percentage of cancer cells, it is critical to determine whether the nanoparticles were able to target CD133+ cells in vivo. The results of the present study demonstrated that CD133-SAL-NPs were able to increase the in vitro therapeutic efficacy of salinomycin in CD133+ ovarian cancer cells. Furthermore, CD133-SAL-NPs preferentially eliminated CD133+ ovarian cancer cells in mice bearing ovarian cancer xenografts.
Although CD133-SAL-NPs exerted superior antitumor efficacy compared with the other treatments assessed by the present study, they were unable to completely eliminate ovarian cancer. There are several reasons that may have contributed to the incomplete elimination of ovarian cancer following CD133-SAL-NP treatment. First, as ovarian cancer possesses distinct CSC subpopulations, the elimination of one subpopulation of CSCs does not result in the elimination of the cancer as a whole (28). CD117 and CD44 are also ovarian CSC markers (5). Thus, nanoparticles conjugated with CD133, CD117 and CD44 may achieve improved therapeutic efficacy for patients with cancer. Second, the CSC phenotype is not stable, as reflected by the ease of conversion from CSCs to non-CSCs (29). Thus, targeting CSCs is insufficient for the complete eradication of cancer. As expected, because the released salinomycin from nanoparticles also targeted non-CSCs, CD133-SAL-NPs were not only able to kill CSCs, but also bulky cancer cells. However, salinomycin is not as potent drug toward non-CSCs as paclitaxel (12). Thus, the therapeutic efficacy of CD133-SAL-NPs may be enhanced in combination with conventional chemotherapy drugs, including paclitaxel.
The anticancer effects of CD133-SAL-NPs were clarified to occur through the following mechanism. First, CD133-SAL-NPs gradually accumulated in ovarian cancer owing to the long circulation time of PEGylated nanoparticles. Following the accumulation of nanoparticles in ovarian cancer, CD133-SAL-NPs were able to specifically bind to and be internalized in CD133+ ovarian CSCs, which resulted in increased cytotoxicity towards CD133+ ovarian CSCs. Concurrently, salinomycin released from CD133-SAL-NPs effectively killed non-CSCs. In contrast, untargeted SAL-NPs may have undergone nonspecific endocytosis and rupture.
To the best of our knowledge, the present study is the first to report the potent activity of salinomycin towards ovarian CSCs. CD133-SAL-NPs were demonstrated to selectively target CD133+ ovarian CSCs. Therefore, CD133 represents a promising target for the delivery of salinomycin to ovarian CSCs, and a feasible mechanism for the inhibition of ovarian cancer through the elimination of CD133+ ovarian CSCs. Collectively, CD133-SAL-NPs represent a promising approach for the treatment of ovarian cancer. As ovarian CSCs serve a pivotal function in the initiation, drug resistance, and metastasis of ovarian cancer, patients with ovarian cancer may benefit from this treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YM and YH did the experiments, analyzed the data, and wrote the manuscript. JD also wrote the manuscript and designed the work. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The experimental protocols were approved by the Institutional Animal Care and Use Committee of Wuhan University (Wuhan, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Authors' information
YM and JD, Department of Obstetrics and Gynecology, Xiangyang No.1 People's Hospital, Hubei University of Medicine, Xiangyang, Hubei 441000, P.R. China; YH, Department of Obstetrics and Gynecology, Renmin Hospital of Wuhan University, Wuhan, Hubei 430060, P.R. China.
References
- 1.Baba T, Convery PA, Matsumura N, Whitaker RS, Kondoh E, Perry T, Huang Z, Bentley RC, Mori S, Fujii S, et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2009;28:209–218. doi: 10.1038/onc.2008.374. [DOI] [PubMed] [Google Scholar]
- 2.Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: Challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 3.Zeimet AG, Reimer D, Sopper S, Boesch M, Martowicz A, Roessler J, Wiedemair AM, Rumpold H, Untergasser G, Concin N, et al. Ovarian cancer stem cells. Neoplasma. 2012;59:747–755. doi: 10.4149/neo_2012_094. [DOI] [PubMed] [Google Scholar]
- 4.Siclari VA, Qin L. Targeting the osteosarcoma cancer stem cell. J Orthop Surg Res. 2010;5:78. doi: 10.1186/1749-799X-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao J, Li W, Guo Y, Feng SS. Nanomedicine strategies for sustained, controlled and targeted treatment of cancer stem cells. Nanomedicine (Lond) 2016;11:3261–3282. doi: 10.2217/nnm-2016-0261. [DOI] [PubMed] [Google Scholar]
- 6.Xie FY, Xu WH, Yin C, Zhang GQ, Zhong YQ, Gao J. Nanomedicine strategies for sustained, controlled, and targeted treatment of cancer stem cells of the digestive system. World J Gastrointest Oncol. 2016;8:735–744. doi: 10.4251/wjgo.v8.i10.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah MM, Landen CN. Ovarian cancer stem cells: Are they real and why are they important? Gynecol Oncol. 2014;132:483–489. doi: 10.1016/j.ygyno.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupia M, Cavallaro U. Ovarian cancer stem cells: Still an elusive entity? Mol Cancer. 2017;16:64. doi: 10.1186/s12943-017-0638-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;21:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue W, Hamaï A, Tonelli G, Bauvy C, Nicolas V, Tharinger H, Codogno P, Mehrpour M. Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy. 2013;9:714–729. doi: 10.4161/auto.23997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Z, Chen D, Xie F, Liu J, Zhang H, Zou H, Yu Y, Chen Y, Sun Z, Wang X, et al. Codelivery of salinomycin and doxorubicin using nanoliposomes for targeting both liver cancer cells and cancer stem cells. Nanomedicine (Lond) 2016;11:2565–2579. doi: 10.2217/nnm-2016-0137. [DOI] [PubMed] [Google Scholar]
- 12.Xie F, Zhang S, Liu J, Gong Z, Yang K, Zhang H, Lu Y, Zou H, Yu Y, Chen Y, et al. Codelivery of salinomycin and chloroquine by liposomes enables synergistic antitumor activity in vitro. Nanomedicine (Lond) 2016;11:1831–1846. doi: 10.2217/nnm-2016-0125. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhang H, Wang X, Wang J, Zhang X, Zhang Q. The eradication of breast cancer and cancer stem cells using octreotide modified paclitaxel active targeting micelles and salinomycin passive targeting micelles. Biomaterials. 2012;33:679–691. doi: 10.1016/j.biomaterials.2011.09.072. [DOI] [PubMed] [Google Scholar]
- 14.Zhao P, Dong S, Bhattacharyya J, Chen M. iTEP nanoparticle-delivered salinomycin displays an enhanced toxicity to cancer stem cells in orthotopic breast tumors. Mol Pharm. 2014;11:2703–2712. doi: 10.1021/mp5002312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J, Gao X, Su L, Xia H, Gu G, Pang Z, Jiang X, Yao L, Chen J, Chen H. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials. 2011;32:8010–8020. doi: 10.1016/j.biomaterials.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Gao J, Lu Y, Kou G, Zhang H, Fan L, Sun Z, Guo Y, Zhong Y. Preparation and characterization of PE38KDEL-loaded anti-HER2 nanoparticles for targeted cancer therapy. J Control Release. 2008;128:209–216. doi: 10.1016/j.jconrel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Manjappa AS, Chaudhari KR, Venkataraju MP, Dantuluri P, Nanda B, Sidda C, Sawant KK, Murthy RS. Antibody derivatization and conjugation strategies: Application in preparation of stealth immunoliposome to target chemotherapeutics to tumor. J Control Release. 2011;150:2–22. doi: 10.1016/j.jconrel.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Liu W, Xia Y, Li W, Sun J, Chen H, Li B, Zhang D, Qian W, Meng Y, et al. The promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomes. Biomaterials. 2011;32:3459–3470. doi: 10.1016/j.biomaterials.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Gao J, Sun J, Li H, Liu W, Zhang Y, Li B, Qian W, Wang H, Chen J, Guo Y. Lyophilized HER2-specific PEGylated immunoliposomes for active siRNA gene silencing. Biomaterials. 2010;31:2655–2664. doi: 10.1016/j.biomaterials.2009.11.112. [DOI] [PubMed] [Google Scholar]
- 20.Nagy I, Caelers A, Monge A, Bonabi S, Huber AM, Bodmer D. NF-kappaB-dependent apoptotic hair cell death in the auditory system. Audiol Neurotol. 2007;12:209–220. doi: 10.1159/000101328. [DOI] [PubMed] [Google Scholar]
- 21.Gao J, Yu Y, Zhang Y, Song J, Chen H, Li W, Qian W, Deng L, Kou G, Chen J, Guo Y. EGFR-specific PEGylated immunoliposomes for active siRNA delivery in hepatocellular carcinoma. Biomaterials. 2012;33:270–282. doi: 10.1016/j.biomaterials.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 22.Kumari A, Yadav SK, Yadav SC. Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf B Biointerfaces. 2010;75:1–18. doi: 10.1016/j.colsurfb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Green MR, Manikhas GM, Orlov S, Afanasyev B, Makhson AM, Bhar P, Hawkins MJ. Abraxane, a novel Cremophor-free, albumin-bound particle form of paclitaxel for the treatment of advanced non-small-cell lung cancer. Ann Oncol. 2006;17:1263–1268. doi: 10.1093/annonc/mdl104. [DOI] [PubMed] [Google Scholar]
- 24.Chumsri S, Burger AM. Cancer stem cell targeted agents: Therapeutic approaches and consequences. Curr Opin Mol Ther. 2008;10:323–333. [PubMed] [Google Scholar]
- 25.Gao J, Chen H, Yu Y, Song J, Song H, Su X, Li W, Tong X, Qian W, Wang H, et al. Inhibition of hepatocellular carcinoma growth using immunoliposomes for co-delivery of adriamycin and ribonucleotide reductase M2 siRNA. Biomaterials. 2013;34:10084–10098. doi: 10.1016/j.biomaterials.2013.08.088. [DOI] [PubMed] [Google Scholar]
- 26.Wang CH, Chiou SH, Chou CP, Chen YC, Huang YJ, Peng CA. Photothermolysis of glioblastoma stem-like cells targeted by carbon nanotubes conjugated with CD133 monoclonal antibody. Nanomedicine. 2011;7:69–79. doi: 10.1016/j.nano.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Shvedova AA, Kagan VE. The role of nanotoxicology in realizing the ‘helping without harm’ paradigm of nanomedicine: Lessons from studies of pulmonary effects of single-walled carbon nanotubes. J Intern Med. 2010;267:106–118. doi: 10.1111/j.1365-2796.2009.02188.x. [DOI] [PubMed] [Google Scholar]
- 28.Visvader JE, Lindeman GJ. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D'Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.