Abstract
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression that target mRNAs for translational repression or cleavage. The present study was conducted to identify differentially expressed miRNAs in primary tumor tissues of rectal carcinoma (RC) that may be associated with heterochrony hepatic metastasis (HHM). Samples were collected exclusively from patients with RC but not colon cancer (CC); Next-generation high-throughput sequencing technology and bioinformatics tools were used to profile and analyze small RNAs and their corresponding targets in primary tumor tissues with HHM (n=2) or without metastases (non-metastatic, NM; n=2). A total of 24 known miRNAs were identified to be differentially expressed (P<0.01; absolute value of log2-fold change ≥1). Hsa-let-7e-5p exhibited the most significant elevation in tissues with HHM (log2-fold change=2.62). By combining online informatics resources and previous mRNA sequencing data, it was identified that 54 validated target genes of let-7e were downregulated in primary tumor tissues with HHM. A number of these target genes have been demonstrated to be directly involved in tumor metastasis (including MYC proto-oncogene, bHLH transcription factor, high-mobility group AT-Hook 2, peptidase inhibitor 3, KIT proto-oncogene receptor tyrosine kinase, Jun proto-oncogene, AP-1 transcription factor subunit and ribonuclease T2), or have physiological associations to immunity (including C-C motif chemokine receptor 4 and cluster of differentiation 40 ligand) and cellular metabolism (including peroxisome proliferator-activated receptor γ, coactivator 1 α). Next, 14 target genes were selected for reverse transcription-quantitative polymerase chain reaction analysis in non-sequenced samples, and the downregulation of 10 target genes in RC samples with HHM was confirmed. In addition, it was demonstrated that hsa-let-7e-5p stimulated colorectal cancer cell migration in vitro. The miRNA hsa-let-7e-5p may serve as a potential biomarker for rectal carcinoma-associated HHM, facilitating the identification of patients with RC who are at risk of developing HHM.
Keywords: microRNA hsa-let-7e-5p, rectal carcinoma, heterochrony hepatic metastasis, next-generation sequencing
Introduction
Colorectal cancer (CRC) is the second most common malignant neoplasm in women and the third in men, with 1.2 million annual incidences worldwide (1). CRC is responsible for 10% of cases of cancer incidence and mortality, worldwide (2). The primary cause for mortality due to colon cancer is liver metastasis, and ~60% of patients may develop metastases (3). The 5-year survival rate of CRC patients is 91% if the carcinoma remains localized at the time of diagnosis; however, this decreases to 11% if distant metastases have occurred (2). Understanding the underlying mechanisms behind CRC tumorigenesis and metastasis will improve clinical treatment. Until recently, CRC has been divided into two categories: Colon carcinoma (CC) and rectal carcinoma (RC) (4). Unfortunately, numerous previous studies have not distinguished between these two distinct forms of carcinoma (5–8).
MicroRNAs (miRNAs/miRs) are evolutionarily conserved ~22-nt-long, non-coding RNAs that bind to the 3′-untranslated regions of mRNA to regulate its expression, thereby inhibiting translation driving the cleavage of target mRNAs (9–11). Previous studies have investigated the association between miRNA polymorphisms and CRC incidence and prognosis, and the possibility of using circulatory or fecal miRNAs as early non-invasive prognosis biomarkers (5,12–15).
In the present study, samples were collected exclusively from patients with RC-positive and CC-negative disease. A next-generation high-throughput sequencing strategy was applied to identify differentially expressed miRNAs in primary tumor tissues of RC with heterochrony hepatic metastases (HHM) or without metastases (non-metastatic, NM). Hsa-let-7e-5p was identified to be the most significantly upregulated in primary tumor tissues with HHM. The potential targets of hsa-let-7e-5p, initially derived from online bioinformatics resources, were additionally verified using RNA sequencing (RNA-seq) data. The majority of target genes have already been identified to be directly involved in tumor metastases. The results of the present study indicate that hsa-let-7e-5p may be used as a prognosis marker to identify patients with RC who may be at risk of metastases.
Patients and methods
Patients and samples
Rectal cancer samples were collected from patients who underwent surgical resection in the Department of Colorectal Surgery, Peking University School of Oncology, Beijing Institute for Cancer Research, Beijing Cancer Hospital (Beijing, China) between February 2007 and November 2010. All patients underwent complete history and physical examination, laboratory evaluation, colonoscopy and biopsy of the lesion, and were diagnosed with rectal carcinoma by pathological diagnosis (hematoxylin-eosin staining and immunohistochemistry). The pre-operative imaging examination and intra-operative probe were used to confirm whether or not synchronous liver metastasis had occurred (16). Briefly, prior to separation and removal of the cancer tissue, a physical examination was conducted to determine whether there were any transferred carcinoma nodules on the surface of the liver or other organs outside of the rectal tissue. Rectal carcinoma patients with concurrent liver metastasis, or in whom liver metastasis developed in the 6 months following surgery, were excluded. According to the guidelines of the National Comprehensive Cancer Network (NCCN), the American Joint Committee on Cancer (AJCC) and College of American Pathologists (17), all patients underwent R0 resection and a minimum of 12 lymph nodes were used to accurately stage the cancer. Patients who had received chemotherapy or radiotherapy prior to surgery were excluded, as were those with other primary malignant tumors, including CC, or those who succumbed to diseases other than RC, or succumbed to RC within 5 years post-surgery (18). In the results of the present study, the accurate postoperative pathological staging information is provided. The American Joint Committee for Cancer (AJCC) seventh edition was used for cancer staging (19). According to this staging, the patients should preoperatively receive chemo- and/or radiotherapy. However, the preoperative radiological staging is not completely in accordance with the postoperative pathological staging. Endoscopic ultrasonography is reported to assess the tumor (T) stage and nodal involvement with 67–97 and 64–88% accuracy, respectively. A meta-analysis of 83 studies demonstrated that computed tomography has 73% accuracy for T stage and 22–73% accuracy for nodal staging. Additionally, the newly endorectal coil magnetic resonance imaging has 71–85% accuracy for prediction of the T stage and 63% for nodal staging of rectal tumors (20,21). Prior to surgery, only preliminary clinical staging of tumors using imaging diagnosis is available. As such, the preoperative evaluation determined that the clinical staging of these patients had not reached the standard of preoperative neoadjuvant chemoradiation; therefore, preoperative chemo- and/or radiotherapy were not administered. Additionally, pathological staging is a key factor of assessing postoperative survival and recurrence for patients with rectal cancer. The AJCC postoperative pathological staging information was summarized to confirm that there were no differences in the pathological staging between the selected patients, ensuring consistency between sequencing samples and comparability between sequencing results (19). HHM was defined as patients who suffered from liver metastases after 6 months of an initial RC diagnosis, and was pathologically confirmed (22). Following this, 84 patients consisted of 51 men (61%) and 33 women (39%) were included and followed up consecutively for 5 years, or until liver metastases appeared. Patients were aged between 38 and 77 years (median, 61 years old). The follow-up and evaluation protocols conformed to the NCCN guidelines (23). The Tumor-Node-Metastasis staging for RC was determined according to the 7th edition of the AJCC Cancer Staging Manual (19). Six cycles of bolus fluorouracil/leucovorin (400 mg/m2/day for fluorouracil, intravenous 200 mg/m2/day for leucovorin, intravenous, 1–5 days/week, every four weeks), were started two weeks following surgery for patients with stage III or stage II disease at high risk of recurrence (tumor perforation, tunica serosa or adjacent organs invasion, venous or lymphatic or peri-neural invasion, poor histopathological differentiation), followed by concurrent 5-FU/radiotherapy (45–50 Gy in 25–28 fractions to the pelvis) (23).
The primary lesions of RC were collected and temporarily stored in liquid nitrogen (−196°C) within 30 min of excision. For long-term storage, the freshly frozen tumor tissues were then stored in the tissue bank (−80°C) in Peking University School of Oncology (Beijing, China) until the end of the follow-up period. The present study was approved by the Medical Ethics Committee of Peking University School of Oncology. Written informed consent was obtained from participants for their clinical records to be used in the present study.
Small RNA isolation and sequencing
Total RNA was extracted from fresh-frozen tissues of patients with RC in the HHM and the NM groups using the RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturers protocol. RNA quality was assessed by 1% agarose gel electrophoresis and by spectrophotometry (260 nm). RNA integrity was assessed using an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). RNAs 18–30-nt in length were isolated and purified from total RNA using Noves 15% TBE-Urea Gel (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Next, the 5′ and 3′ adaptors (Beijing Genomics Institute, Shenzhen, China) for additional sequencing were ligated and the small RNA were reverse transcribed. Finally, following an amplification procedure, the products were sequenced using an Illumina Genome Analyzer (BGI Biotechnology, Cambridge, MA, USA), as previously described (24).
Identification of differentially expressed miRNAs
The analysis procedure for differentially expressed miRNAs has been described previously (25). Firstly, the raw sequencing data were analyzed using adaptor sequences filtering and base calling using Phred (http://www.phrap.com/). Secondly, the overall length of RNAs distribution was calculated and the sequences were mapped onto the human genome using the program Short Oligo nucleotide Analysis Package v2.04 (26). Next, the differences in small RNA sequences between HHM and NM groups were detected. Different types of RNA (small nuclear RNA, tRNA, rRNA and small nucleolar RNA, hereafter referred to as rRNAetc) were identified by blasting with the database of NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and Rfam (version 10.1; http://rfam.xfam.org/). Finally, the small RNAs were compared with the known miRNAs using miRBase 20.0 (http://www.mirbase.org/). The RNAs were marked as ‘unann’ if they were unable to be mapped to any known databases.
Differential miRNA expression levels between NM and HHM were analyzed according to a previously described method (27). In the present study, pairwise comparisons were conducted to identify the differences in miRNA expression between the HHM and NM groups. The expression difference was assessed using the fold change of reads per kilobase of transcript per million mapped reads, and the P-value according to the following formulas: Fold change=log2 (HHM/NM)
Where fold change represents the fold change of miRNA expression in the HHM group relative to the NM group, x indicates the observed the number of reads for the miRNA in one library, y represents the observed number of reads for the miRNA in another library, and N1 and N2 are the total reads for the two libraries, respectively. In the present study, a miRNA was considered to be significantly differentially expressed if the absolute value of the fold change was ≥1 and P<0.01.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
For RT-qPCR of mRNA, total RNA from rectal carcinoma tissues were extracted using RNAiso plus (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturers protocol, and then subjected to RT with random hexamers using the RevertAid First Strand cDNA Synthesis System (Thermo Fisher Scientific, Inc.) to obtain cDNA. qPCR was performed using an UltraSYBR Mixture (with High ROX) (CW Biotech. Co., Ltd., Beijing, China; http://www.cwbiotech.com/) on the LightCycler 480 II (Roche Applied Science, Penzberg, Germany). The thermocycling conditions were as follows: Polymerase activation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. The 2−ΔΔCq method was used, where the Cq value of one target gene was compared to the Cq value of GAPDH internal control gene, in HHM and NM groups (25). Primers are presented in Table I.
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| Target genes | NCBI Gene ID | Direction | Sequence (5′-3′) |
|---|---|---|---|
| NKX2-1 | 7080 | Forward | AGCACACGACTCCGTTCTC |
| Reverse | GCCCACTTTCTTGTAGCTTTCC | ||
| MSTN | 2660 | Forward | GACGATTATCACGCTACAACGG |
| Reverse | TCCATAGTTGGGCCTTTACTACT | ||
| CCR4 | 1233 | Forward | AGAAGGCATCAAGGCATTTGG |
| Reverse | ACACATCAGTCATGGACCTGAG | ||
| PRTN3 | 5657 | Forward | AACTACGACGCGGAGAACAAA |
| Reverse | CGAGGGACGAAAGTGCAAATG | ||
| TREH | 11181 | Forward | TGGGCTTGCTCATTCAGTCA |
| Reverse | TCCCCGTGGCAGTAAATCTC | ||
| GHR | 2690 | Forward | CCATTGCCCTCAACTGGACTT |
| Reverse | AATATCTGCATTGCGTGGTGC | ||
| NR1I2 | 8856 | Forward | GCCCATGCTGAAATTCCACTA |
| Reverse | GCCGATTGCATTCAATGTAGGA | ||
| PROM1 | 8842 | Forward | AGTCGGAAACTGGCAGATAGC |
| Reverse | GGTAGTGTTGTACTGGGCCAAT | ||
| PPARGC1A | 10891 | Forward | TGAAGACGGATTGCCCTCATT |
| Reverse | GCTGGTGCCAGTAAGAGCTT | ||
| CD40LG | 959 | Forward | GAGCAACAACTTGGTAACCCT |
| Reverse | GGCTGGCTATAAATGGAGCTTG | ||
| PI3 | 5266 | Forward | CACGGGAGTTCCTGTTAAAGG |
| Reverse | TCTTTCAAGCAGCGGTTAGGG | ||
| KIT | 3815 | Forward | TGCTCTGCTTCTGTACTGCC |
| Reverse | GCCTTACATTCAACCGTGCC | ||
| MYC | 4609 | Forward | CATCAGCACAACTACGCAGC |
| Reverse | GCTGGTGCATTTTCGGTTGT | ||
| JUN | 3725 | Forward | GTGCCGAAAAAGGAAGCTGG |
| Reverse | CTGCGTTAGCATGAGTTGGC | ||
| GAPDH | 2597 | Forward | GGAGCGAGATCCCTCCAAAAT |
| Reverse | GGCTGTTGTCATACTTCTCATGG | ||
| U6 | 26827 | Forward | AATTGGAACGATACAGAGAAGATT |
| Reverse | TATGGAACGCTTCACGAATTTG |
NCBI, National Center for Biotechnology Information; NKX2-1, NK2 homeobox 1; MSTN, myostatin; CCR4, C-C motif chemokine receptor 4; PRTN3, proteinase 3; TREH, trehalase; GHR, growth hormone receptor; NR1I2, nuclear receptor subfamily 1 group I member 2; PROM1, prominin 1; PPARGC1A, PPARG coactivator 1 α; CD40LG, CD40 ligand; PI3, peptidase inhibitor 3; KIT, KIT proto-oncogene receptor tyrosine kinase; MYC, MYC proto-oncogene, bHLH transcription factor; JUN, Jun proto-oncogene, AP-1 transcription factor subunit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
For the miRNA RT-qPCR, a miRNA first-strand cDNA synthesis kit (cat. no. KR211; Tiangen Biotech Co., Ltd., Beijing, China) was first used to complete the reverse transcription of let-7e-5p from total RNA of rectal carcinoma tissues, including miRNA. Total RNA were extracted using RNAiso plus buffer (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. Briefly, miRNAs were ligated to poly(A) and reverse transcribed with a poly(T) adapter primer. The thermocycling conditions were 42°C for 120 min and 95°C for 3 min. Next, the let-7e-5p-specific forward and reverse primers were used to perform qPCR with miRcute miRNA qPCR SYBR Green detection kit (cat. no. FP411; Tiangen Biotech Co., Ltd.,) according to the manufacturers protocol. The let-7e-5p-specific forward primer was purchased from Tiangen Biotech Co., Ltd (cat. no. CD201-0002). The 2−ΔΔCq method was (28), where the Cq value of miRNA was compared to the Cq value of U6 internal control in HHM and NM groups.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
GO annotation for let-7e-5p targets was performed by mapping all of the targets to the GO database (http://www.geneontology.org/). Next, GO functional classification for annotated targets was processed using WEGO software (29). Finally, GO enrichment analysis was conducted using the hypergeometric test to identify significantly enriched GO terms in targets compared with the whole genome background. The formula used was as follows:
Where N is the number of all genes with GO annotations, n is the number of targets in N, M is the number of all genes that are annotated to the certain GO terms, and m is the number of targets in M. The present study adopted a Bonferroni corrected P-value of ≤0.05 as a threshold. Those terms fulfilling this condition are defined as significantly enriched GO terms in DEGs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database is the major public pathway-related database (30). KEGG pathway enrichment analysis identifies significantly enriched metabolic pathways or signal transduction pathways in targets compared with the whole genome background. The calculation formula and analysis procedure were the same as those used in GO analysis.
In vitro scratch migration assays
Colorectal adenocarcinoma Caco-2 cells were provided by the Stem Cell Bank, Chinese Academy of Sciences (http://www.cellbank.org.cn/). The base medium for this cell line was American Type Culture Collection-formulated Eagle's Minimum Essential Medium (cat. no. 30-2003; American Type Culture Collection, Manassas, VA, USA). The complete growth medium was generated by adding fetal bovine serum (TransGen Biotech Co., Ltd, Beijing, China), giving a final concentration of 20%, to the base medium. The culture conditions were 95% air, 5% CO2 and 37°C. Pcdna3.1-enhanced green fluorescent protein (EGFP)-hsa-pre-let-7e-5p and Pcdna3.1-EGFP-hsa-let-7e-5p-sponge plasmids were constructed by ligating the hsa-pre-let-7e-5p and hsa-let-7e-5p-sponge (Sangon Biotech Co., Ltd, Shanghai, China) downstream of Pcdna3.1-EGFP (Miaoling Biotech Co., Ltd, Wuhan, China) using T4 DNA ligase (Takara Biotechnology Co., Ltd.). Following transfection with Pcdna3.1-EGFP-hsa-pre-let-7e-5p, Pcdna3.1-EGFP-hsa-let-7e-5p-sponge or Pcdna3.1-EGFP using Lipofectamine 3000 (Thermo Fisher Scientific Inc., Waltham, MA, USA), cells were grown in tissue culture dishes at 37°C for 12 h, and scratches were made using a 10 µl pipette tip. A total of 10 different areas were marked on the bottom of the chamber, and images of these spots were captured using a fluorescent microscope (magnification, ×10) at time intervals (0, 24 and 48 h following scratching). Migration of the cells was measured using the ImageJ V1.5 software package (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
The differences in mRNA and miRNA levels between paired samples were determined by the Wilcoxon matched-pairs test using GraphPad Prism V6.0 (GraphPad Software, Inc., La Jolla, CA USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics and samples grouping
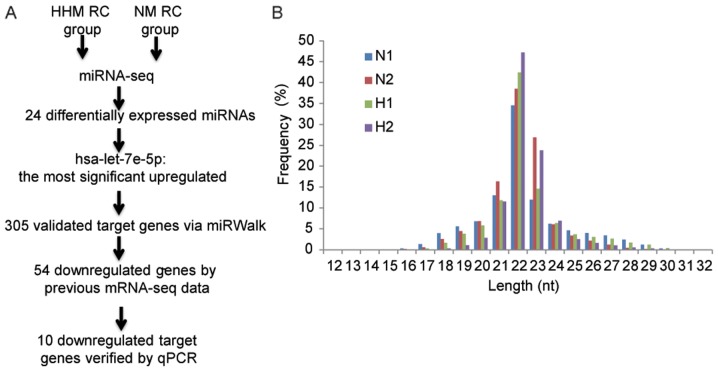
A total of 84 patients with RC were followed from the day 1 post-surgery, with a median time of 61 months and the proportion of patients who adhered to follow-up was 98.8% (83/84). The follow-up data indicated that 64 patients exhibited NM, 6 patients developed local recurrence and 13 exhibited HHM. From the NM and HHM groups, 5 primary RC tumor samples were randomly selected for RNA extraction. Following RNA quality and integrity assessment, 2 NM samples and 3 HHM samples were selected for additional sequencing (Fig. 1A).
Figure 1.
miRNA sequencing of RC samples. (A) The workflow for the present study. (B) The distribution of length of the small RNA library in the NM RC and HHM RC samples. miRNA, microRNA; nt, nucleotides; RC, rectal carcinoma; NM, non-metastasis; HHM, heterochrony hepatic metastasis; N1 and N2, NM RC samples; H1 and H2, HHM samples; seq, sequencing.
Sequencing and analysis of small RNAs from NM and HHM RC
To investigate the role of miRNAs in rectal carcinoma with HHM, miRNA expression profiles in primary tumor tissues of rectal carcinoma with HHM (n=3) and NM (n=2) were determined by next-generation high-throughput sequencing technology (Table II). Small RNAs length distribution in 4 samples was 22 nt, with the exception of one HHM sample (Fig. 1B). It is possible that the low quality of miRNA library construction of this HHM sample resulted in abnormal miRNA length distribution. Therefore, only two HHM samples were used for subsequent analysis. For each sample, 18.3–23.6 million reads were obtained. Following the elimination of the adaptor sequence reads and sequences <18 nt, each sample contained >97% (~17 million) clean reads 18–30 nt in length. In total, >70% (74.7–79.0%) clean reads from each sample were mapped to genomic regions, resulting in 676,280 (36.44%), 486,314 (40.94%), 434,966 (31.8%) and 4,251,607 (38.03%) that matched with the small RNA region for tissues with HHM and NM respectively.
Table II.
Clinical information of the patients with rectal cancer.
| A, Sequenced samples | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Metastasis | Sex | Age, years | Tumor location | Tumor size, cm | Pathological type | Vascular invasion | pTNM stagea | Preoperative serum CEA levels (μg/l) | Ki67, %c | Postoperative HHM time, months |
| N1 | NM | F | 58 | Upper | 5.1 | Ulcer type, moderately differentiated adenocarcinoma | − | T4aN0M0, IIb | 5.2 | 45–60 | − |
| N2 | NM | M | 66 | Upper | 4.5 | Ulcer type, moderately or poorly differentiated adenocarcinoma | + | T4aN2bM0, IIIC | 2.8 | <25 | − |
| H1 | HHM | M | 60 | Upper | 6.0 | Ulcer type, moderately or poorly differentiated adenocarcinoma | + | T4bN1M0, IIIC | 2.9 | 50–75 | 48 |
| H2 | HHM | M | 38 | Upper | 5.0 | Ulcer type, moderately or poorly differentiated adenocarcinoma, mucinous partially | + | T4aN1M0, IIIB | 17.0 | 50 | 7 |
| H3b | HHM | F | 77 | Middle | 5.0 | Ulcer type, moderately differentiated adenocarcinoma | − | T3N0M0, IIa | 5.9 | 60 | 15 |
| B, Non-sequenced samples | |||||||||||
| N3 | NM | M | 62 | Middle | 4.3 | Ulcer type, moderately differentiated adenocarcinoma | − | T3N0M0, IIa | 4.7 | 35 | − |
| N4 | NM | M | 73 | Middle | 5.0 | Ulcer type, moderately differentiated adenocarcinoma | + | T3N1M0, IIIB | 16.6 | <25 | − |
| H4 | HHM | M | 61 | Middle | 5.0 | Ulcer type, moderately differentiated adenocarcinoma | + | T4bN2aM0, IIIC | 8.4 | 10 | 10 |
| H5 | HHM | F | 46 | Middle | 5.0 | Ulcer type, moderately differentiated adenocarcinoma | − | T3N0M0, IIa | 3.7 | 65 | 63 |
The TNM stage and histological grading were determined according to the Union for International Cancer Control criteria. The histological classification was based on the WHO criteria. The tumor locations were categorized according to the standard guidelines for clinical practice. Lower rectal cancer, distance between the bottom edge of tumor to anal verge within 3–6 cm; middle rectal cancer, 6–10 cm; upper rectal cancer, 10–15 cm.
The small RNAs length distribution of the H3 sample did not peak at 22 nucleotides, and therefore H3 was not used for additional analysis.
The proportion of Ki67 positive cells. F, female; M, male; NM, non-metastasis; HHM, heterochrony hepatic metastasis; CEA, carcinoembryonic antigen; pTNM, pathological Tumor Node Metastasis; Ki67, antigen Ki67.
Next, the clean small RNA reads were analyzed by mapping them to the non-coding RNAs in the GenBank, Rfam 10.1 and miRBase 20.0 databases. All clean reads were divided into known miRNAs and all other rRNA (Table III), and the rRNAetc fractions were removed from all the samples. The number of unann small RNAs was generally high when it was calculated from the unique reads small RNAs (Table III). However, when it was calculated from the total small RNAs, the number of unann small RNAs was ~20% of total reads (Table III), indicating that the majority of the total small RNAs were miRNAs. Briefly, 8,771,735 (38.57% of small RNA reads) and 9,081,651 (47.47% of small RNA reads) tags from the patients with NM, and 8,506,675 (47.82% of small RNA reads) and 10,871,492 (63.29% of small RNA reads) tags from the patients with HHM were matched to hairpin precursors of known miRNAs. Examination of these reads in the small RNA libraries indicated the presence of a high percentage of unann small RNAs, ~20% for all samples (Table III).
Table III.
Composition of sRNAs among different categories.
| A, N1 | ||||
|---|---|---|---|---|
| Category | Total reads | % | Unique reads | % |
| Total sRNAs | 22,740,286 | 100 | 1,855,776 | 100 |
| sRNAs match hairpin | 8,771,735 | 38.57 | 5,687 | 0.31 |
| rRNAetc | 9,975,175 | 43.87 | 878,176 | 47.32 |
| Unann | 3,993,376 | 17.56 | 971,913 | 52.37 |
| B, N2 | ||||
| Total sRNAs | 19,130,301 | 100 | 1,187,729 | 100 |
| sRNAs match hairpin | 9,081,651 | 47.47 | 4,910 | 0.41 |
| rRNAetc | 6,422,337 | 33.57 | 632,232 | 53.23 |
| Unann | 3,626,313 | 18.96 | 550,587 | 46.36 |
| C, H1 | ||||
| Total sRNAs | 17,788,876 | 100 | 1,366,299 | 100 |
| sRNAs match hairpin | 8,506,675 | 47.82 | 4,835 | 0.35 |
| rRNAetc | 5,580,275 | 31.37 | 588,867 | 43.10 |
| Unann | 3,701,926 | 20.81 | 772,597 | 56.55 |
| D, H2 | ||||
| Total sRNAs | 17,177,276 | 100 | 661,686 | 100 |
| sRNAs match hairpin | 10,871,492 | 63.29 | 4,583 | 0.69 |
| rRNAetc | 2,466,095 | 14.36 | 313,039 | 47.31 |
| Unann | 3,839,689 | 22.35 | 344,064 | 52.00 |
RC, rectal carcinoma; unann, unannotated small RNAs; sRNAs, small RNAs; N1 and N2, RC samples without metastasis; H1 and H2, RC samples with heterochrony hepatic metastasis; rRNAetc, small nuclear RNA, tRNA, rRNA and small nucleolar RNA.
miRNAs are differentially expressed in primary tumor tissues of rectal carcinoma with and without HHM
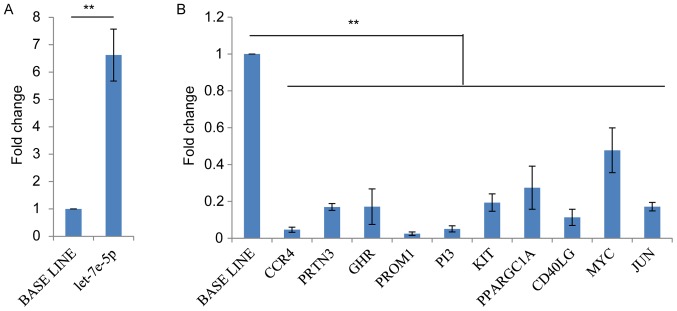
Pair-wise intersection analysis demonstrated that >90% of small RNAs were commonly expressed among all samples. Differential expression analysis was performed for the HHM and NM groups. In total, 24 significantly differentially expressed miRNAs (P<0.01; the absolute value of log2-fold change >1) were identified (Table IV). Among these differentially expressed miRNAs, 9 upregulated miRNAs, with log2-fold change >1.5 in the HHM samples compared with the NM samples, were identified. Concurrently, 15 downregulated miRNAs exhibited log2-fold change ~2 in the HHM samples. Among the upregulated miRNAs, hsa-let-7e-5p exhibited the highest normalized expression level in the tissues with (H-std=7,774.26±1,434.92 reads) and without HHM (N-std=1,201.21±29.95 reads), and exhibited a log2-fold change of 2.62 in tissues with HHM (Table IV). Expression of hsa-miR-224-5p was also upregulated, with a high normalized expression level (H-std=175.01±11.59 reads; N-std=52.21±5.14 reads) (Table IV). Conversely, the normalized expression levels of all the downregulated miRNAs were low compared with upregulation miRNAs (all <100 reads; Table IV), with the exception of hsa-miR-424-5p, with 159.04±26.06 reads for tissues without HHM. RT-qPCR confirmed that hsa-let-7e-5p was upregulated >5-fold in the HHM samples (Fig. 2A). Consequently, all additional analysis focused on hsa-let-7e-5p.
Table IV.
Significantly differentially expressed microRNAs between RC samples with HHM and without metastasis.
| miRNA | N-stha | H-stha | Fold-change (log2 H/N) | P-value |
|---|---|---|---|---|
| hsa-let-7e-5p | 1,201.20±29.95 | 7,774.26±1,434.92 | 2.62±0.28 | <0.01 |
| hsa-miR-125a-3p | 0.47±0.03 | 1.83±0.15 | 1.94±0.15 | <0.01 |
| hsa-miR-1307-5p | 70.80±15.95 | 12.90±3.82 | −2.55±0.58 | <0.01 |
| hsa-miR-188-5p | 3.41±0.97 | 0.45±0.13 | −2.90±0.63 | <0.01 |
| hsa-miR-224-5p | 52.21±5.14 | 175.01±11.59 | 1.76±0.17 | <0.01 |
| hsa-miR-23a-5p | 3.41±0.70 | 19.01±2.17 | 2.54±0.35 | <0.01 |
| hsa-miR-362-3p | 11.59±0.93 | 2.56±0.34 | −2.20±0.23 | <0.01 |
| hsa-miR-3648 | 5.36±0.44 | 1.16±0.34 | −2.40±0.47 | <0.01 |
| hsa-miR-3651 | 2.09±0.06 | 0.48±0.21 | −2.74±0.83 | <0.01 |
| hsa-miR-3687 | 6.67±0.37 | 1.45±0.33 | −2.32±0.36 | <0.01 |
| hsa-miR-378d | 2.63±0.46 | 0.51±0.16 | −2.55±0.57 | <0.01 |
| hsa-miR-424-5p | 159.04±26.06 | 35.56±6.12 | −2.17±0.35 | <0.01 |
| hsa-miR-432-5p | 16.75±0.02 | 55.15±5.13 | 1.70±0.14 | <0.01 |
| hsa-miR-4426 | 1.16±0.67 | 0.31±0.08 | −2.04±0.40 | 0.01 |
| hsa-miR-4454 | 11.68±1.76 | 2.39±0.34 | −2.28±0.31 | <0.01 |
| hsa-miR-449a | 0.34±0.02 | 2.54±0.82 | 2.65±0.53 | <0.01 |
| hsa-miR-449c-5p | 0.79±0.21 | 5.71±0.34 | 3.00±0.41 | <0.01 |
| hsa-miR-493-5p | 1.81±0.20 | 5.12±0.07 | 1.53±0.16 | <0.01 |
| hsa-miR-509-3-5p | 3.81±0.76 | 0.52±0.07 | −2.84±0.36 | <0.01 |
| hsa-miR-509-3p | 2.26±0.39 | 0.23±0.04 | −3.35±0.23 | <0.01 |
| hsa-miR-556-3p | 3.16±0.53 | 0.97±0.06 | −1.65±0.26 | <0.01 |
| hsa-miR-652-5p | 4.13±0.30 | 1.24±0.19 | −1.79±0.26 | <0.01 |
| hsa-miR-7704 | 2.85±0.56 | 0.45±0.23 | −3.60±1.15 | <0.01 |
| hsa-miR-99b-5p | 22.46±0.68 | 69.46±1.54 | 1.63±0.05 | <0.01 |
RC, renal carcinoma; hsa, homo sapiens; NM, non-metastasis; HHM, heterochrony hepatic metastasis; N, NM RC; H, HHM RC.
Std refers to the normalized miRNAs levels.
Figure 2.
RT-qPCR analysis results of hsa-let-7e and its targets. (A) RT-qPCR validation of hsa-let-7e in the sequenced samples. (B) RT-qPCR analysis of 10 hsa-let-7e targets in the non-sequenced samples. **P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; has, Homo sapiens.
Target analysis of hsa-let-7e-5p
To elucidate the potential biological mechanisms of hsa-let-7e-5p in promoting HHM, its potential targets were investigated, which had been validated previously using the miRWalk database (31,32). hsa-let-7e was identified to have 305 validated target genes (data not shown). By examining the mRNA-seq data of the present study (the transcriptome of 5 HHM and 5 NM RC samples), it was revealed that the expression of 54 of these validated target genes was downregulated in tissues with HHM. A number of these 54 target genes have been demonstrated to be directly involved in tumor metastases, including MYC proto-oncogene, bHLH transcription factor (MYC), high-mobility group AT-Hook 2, peptidase inhibitor 3 (PI3), KIT proto-oncogene receptor tyrosine kinase (KIT), Jun proto-oncogene, AP-1 transcription factor subunit (JUN) and ribonuclease T2 (33–36), or have physiological associations with immunity, including C-C motif chemokine receptor 4 (CCR4) (37), cluster of differentiation 40 ligand (CD40LG) (38), and cellular metabolism, for example peroxisome proliferator-activated receptor γ, coactivator 1 α (PPARGC1A) (39). Among the 54 target genes, 12 exhibited a log2-fold reduction >2.0 in gene expression: NK2 homeobox 1; myostatin; CCR4, proteinase 3, trehalase, growth hormone receptor, nuclear receptor subfamily 1 group I member 2, prominin 1, PPARGC1A, CD40LG, PI3 and KIT (Table V). Next, the expression levels of these 12 targets plus MYC (1.8 log2-fold change) and JUN (1.2 log2-fold change) were compared between 2 additional pairs of HHM and NM RC samples (non-sequenced) (Table II). The results of RT-qPCR indicated that the log2-fold change of 10 out of the 14 target genes were consistent with the results derived from the sequenced samples (Fig. 2B).
Table V.
Downregulated known validated hsa-let-7e-5p target genes from RNA sequencing results.
| Gene name | logN1 vs. H1 | logN1 vs. H2 | logN2 vs. H1 | logN2 vs. H2 | Average |
|---|---|---|---|---|---|
| NKX 20.0011 | −12.47 | −12.47 | −16.01 | −16.01 | −14.24 |
| MSTN | −8.21 | −8.21 | −8.01 | −8.01 | −8.11 |
| CCR4 | −8.39 | −1.54 | −9.20 | −2.34 | −5.37 |
| PRTN3 | −8.12 | −0.54 | −7.92 | −0.34 | −4.23 |
| TREH | −3.22 | −2.86 | −4.16 | −3.80 | −3.51 |
| GHR | −3.16 | −4.38 | −1.12 | −2.34 | −2.75 |
| NR1I2 | −0.80 | −3.64 | −1.60 | −4.44 | −2.62 |
| PROM1 | −3.31 | −1.86 | −3.18 | −1.74 | −2.52 |
| PPARGC1A | −2.24 | −4.20 | −0.84 | −2.80 | −2.52 |
| CD40LG | −3.48 | −2.13 | −2.29 | −0.93 | −2.21 |
| PI3 | −3.70 | −2.37 | −1.97 | −0.64 | −2.17 |
| KIT | −3.12 | −2.18 | −2.02 | −1.08 | −2.10 |
| UHRF2 | −1.19 | −2.47 | −1.12 | −2.40 | −1.79 |
| MYC | −1.15 | −2.14 | −1.36 | −2.34 | −1.75 |
| ZNF167 | −2.04 | −0.30 | −2.84 | −1.10 | −1.57 |
| SLC25A10 | −1.69 | −1.79 | −1.24 | −1.34 | −1.51 |
| CSAD | −0.25 | −1.39 | −1.57 | −2.72 | −1.48 |
| TRAT1 | −0.90 | −0.54 | −2.29 | −1.93 | −1.41 |
| HMGA2 | −2.62 | −1.90 | −0.92 | −0.20 | −1.41 |
| NOL3 | −1.09 | −0.39 | −2.32 | −1.62 | −1.35 |
| JUN | −0.59 | −1.26 | −1.08 | −1.75 | −1.17 |
| DNMT3A | −0.82 | −1.10 | −1.24 | −1.52 | −1.17 |
| RPE | −0.72 | −1.61 | −0.66 | −1.54 | −1.13 |
| TUT1 | −1.56 | −0.34 | −1.84 | −0.62 | −1.09 |
| CYCS | −0.78 | −2.04 | −0.13 | −1.39 | −1.09 |
| RNASET2 | −0.50 | −0.58 | −1.58 | −1.66 | −1.08 |
| CCNT2 | −0.26 | −0.95 | −0.95 | −1.64 | −0.95 |
| RABEPK | −1.41 | −1.50 | −0.32 | −0.40 | −0.91 |
| RPIA | −0.20 | −1.44 | −0.37 | −1.60 | −0.90 |
| IFI44 | −1.66 | −1.09 | −0.70 | −0.14 | −0.90 |
| NCALD | −0.46 | −1.13 | −0.58 | −1.24 | −0.85 |
| DICER1 | −0.72 | −0.58 | −1.06 | −0.92 | −0.82 |
| FASN | −1.57 | −1.09 | −0.54 | −0.05 | −0.81 |
| HNF4A | −1.26 | −0.37 | −1.25 | −0.36 | −0.81 |
| TRIM17 | −1.22 | −0.86 | −0.70 | −0.34 | −0.78 |
| POLD3 | −1.12 | −0.85 | −0.67 | −0.39 | −0.76 |
| NCOA3 | −1.25 | −0.31 | −1.19 | −0.25 | −0.75 |
| VDR | −0.50 | −1.22 | −0.28 | −0.99 | −0.75 |
| SOCS4 | −0.73 | −0.75 | −0.69 | −0.71 | −0.72 |
| COX8A | −1.04 | −1.27 | −0.07 | −0.30 | −0.67 |
| CNN3 | −0.53 | −0.52 | −0.80 | −0.80 | −0.66 |
| CD46 | −0.30 | −1.06 | −0.23 | −0.99 | −0.64 |
| AMACR | −0.11 | −0.20 | −1.09 | −1.18 | −0.64 |
| CHEK1 | −0.55 | −0.77 | −0.38 | −0.61 | −0.58 |
| HMGA1 | −0.30 | −1.02 | −0.12 | −0.84 | −0.57 |
| EIF2C4 | −0.22 | −0.26 | −0.85 | −0.88 | −0.55 |
| HNRNPA1 | −0.11 | −0.69 | −0.25 | −0.83 | −0.47 |
| MEST | −0.11 | −0.29 | −0.62 | −0.80 | −0.45 |
| TRIM32 | −0.43 | −0.62 | −0.29 | −0.48 | −0.45 |
| CCNG1 | −0.05 | −0.67 | −0.12 | −0.74 | −0.39 |
| PSMA7 | −0.44 | −0.72 | −0.07 | −0.35 | −0.39 |
| DYRK2 | −0.14 | −0.35 | −0.34 | −0.55 | −0.35 |
| CASP8 | −0.10 | −0.48 | −0.21 | −0.59 | −0.35 |
| NDUFA2 | −0.08 | −0.38 | −0.07 | −0.38 | −0.23 |
N1 and N2 indicate non-metastasis samples, H1 and H2 indicate heterochrony hepatic metastasis samples.
Gene Ontology (GO) and KEGG analysis of target genes
GO analysis was performed on the 54 validated targets of hsa-let-7e-5p. The target genes were categorized into 18 biological processes, nine cellular components and seven molecular functions. Subsequent analysis indicated that the identified hsa-let-7e-5p targets were enriched in 18 KEGG pathways. Notably, targets were identified to participate in the transforming growth factor-β signaling pathway, the p53 signaling pathway, melanogenesis, Human T-lymphotropic virus-I infection, Epstein-Barr virus infection and pathways in cancer (Table VI), indicating that hsa-let-7e-5p serves a notable role in tumor metastases.
Table VI.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of 54 hsa-let-7e-5p target genes.
| Significant gene symbols | Pathway ID | Pathway name | P-valuea | Q-valueb |
|---|---|---|---|---|
| MYC | ko04350 | TGF-β signaling pathway | 0.0002 | 0.0161 |
| CCNG1 | ko04115 | p53 signaling pathway | 0.0006 | 0.0108 |
| KIT | ko04916 | Melanogenesis | <0.0001 | 0.0035 |
| MYC, JUN, POLD3 | ko05166 | HTLV–I infection | <0.0001 | 0.0277 |
| MYC | ko05169 | Epstein-Barr virus infection | <0.0001 | 0.0342 |
| KIT, MYC, JUN | ko05200 | Pathways in cancer | <0.0001 | 0.0695 |
P-value generated using the hypergeometric test.
Q≤0.05 suggests that this pathway is significantly enriched. Hsa, Homo sapiens; MYC, MYC proto-oncogene, bHLH transcription factor; KIT, KIT proto-oncogene receptor tyrosine kinase; JUN, Jun proto-oncogene, AP-1 transcription factor subunit; CCNG1, cyclin G1; POLD3, DNA polymerase ∆3, accessory subunit; TGF-β, transforming growth factor β; p53, tumor protein; HTLV-I, human T-lymphotropic virus 1.
Overexpression of hsa-let-7e-5p promotes tumor cell migration while knockdown of hsa-let-7e-5p suppresses migration
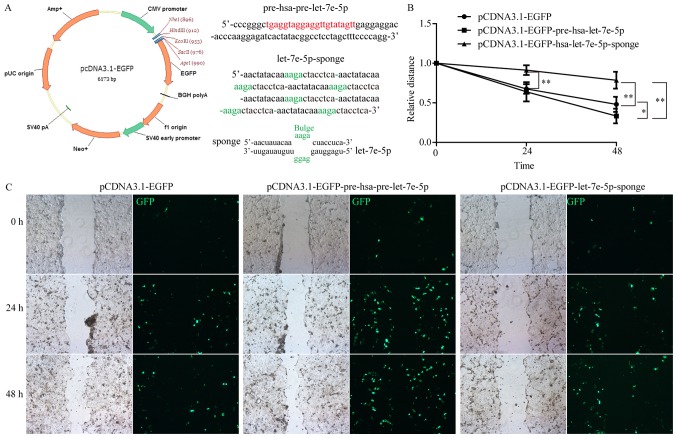
To investigate the potential mechanism of hsa-let-7e-5p in promoting RC progression, Caco-2 cells were transfected with pCDN3.1-EGFP-pre-hsa-let-7e-5p or pCDN3.1-EGFP-hsa-let-7e-5p-sponge plasmids, to determine the effect of hsa-let-7e-5p on cell growth in vitro (Fig. 3A). Elevated hsa-let-7e-5p levels resulted in increased cellular migration in culture, whereas inhibition of hsa-let-7e-5p expression significantly suppressed tumor cell mobility (Fig. 3B). Stable expression of pre-hsa-let-7e-5p or hsa-let-7e-5p-sponge was confirmed by the presence of a GFP signal (Fig. 3C). These results indicated that hsa-let-7e-5p stimulated colorectal cancer cell spreading, indicating that it had a potential role in cancer metastasis.
Figure 3.
Effects of hsa-let-7e-5p overexpression or inhibition on Caco-2 cell migration in vitro. (A) Construction of pCDNA3.1-hsa-pre-let-7e-5p and pCDNA3.1-hsa-let-7e-5p-sponge plasmids. A total of 6 sponge repeats were used for the construction. The red letters indicate mature let-7e-5p sequence, and the green letters indicate bulge region of let-7e-5p-sponge. (B) The effect of let-7e-5p overexpression and inhibition increased and decreased the relative distance of Caco-2 cells migration, respectively. *P<0.05, **P<0.01. (C) Representative images of the wound-healing assay performed in Caco-2 cells with let-7e-5p overexpression or inhibition. The images were captured at 0, 24 and 48 h post-wounding. Data are presented as the mean ± standard deviation of 3 independent experiments. EGFP, enhanced green fluorescent protein.
Discussion
A number of studies have demonstrated that miRNAs serve notable roles in tumor invasion and metastases (5,13,14,40,41). In the present study, miRNA profiles of primary focal tumor tissues from two patients with HHM RC and two patients with NM RC were compared. Using high-throughput sequencing, a small number of differentially expressed miRNAs were identified. Expression of hsa-let-7e-5p was markedly upregulated in HHM RC. Subsequent assessment of the expression of hsa-let-7e-5p target genes implicated that it may be a prognostic biomarker for RC with HHM. However, the small number of sequenced specimens (n=4) is a limitation, and therefore the present data are largely descriptive and should be evaluated in a higher number of cases.
Conversely, the role of hsa-let-7 in cancer development has been controversial. For example, certain studies have demonstrated that let-7f inhibits tumor invasion and metastasis in human gastric cancer, whereas let-7a, let7b and let-7g suppress breast cancer cell migration and invasion (40,42). On the contrary, a previous study indicated that the let-7 miRNA family is secreted from a metastatic gastric cancer cell line into the extracellular environment (43). In addition, a novel let-7-regulated transcriptional factor, transcription regulator factor BACH1, induces the expression of matrix metalloproteinase 1 and promotes metastasis in breast cancer (44).
In the present study, MYC was identified as a target of let-7e that may participate in HHM RC. In previous studies, elevation of MYC has been detected in a variety of human tumors (45–47). Overexpression of MYC may transform cells in culture and elicit malignant tumors in experimental animals (48). However, certain previous studies provided paradoxical data; that this powerful oncogene may also act as a suppressor of cell motility, invasiveness and metastases (33). MYC may suppress metastasis by directly silencing the transcription of integrin proteins (33). These data uncovered an unexpected function of MYC, which provides an explanation for the current controversy on the role of MYC in metastasis (33). In the present study, MYC was downregulated in the HHM RC, indicating a metastasis-suppressing role for MYC.
c-Kit is an additional notable let-7e target that may participate in HHM RC. c-Kit expression is suppressed in colon cancer tissues, contributing to L1-mediated metastases (34). The L1 is a transmembrane neural cell adhesion receptor in human colorectal cancer and promotes liver metastases (34). Although c-Kit upregulation inhibits the metastases of L1-expressing colorectal cancer cells, it also enhances colorectal cancer cell tumorigenesis and proliferation, which indicates that c-Kit may mediate separate pathways in metastases (34).
By contrast, other let-7e candidate targets identified in the present study were demonstrated to inhibit cancer metastasis. For example, cyclin-dependent kinase inhibitor 1B suppresses the growth, migration and metastasis of mouse embryonic fibroblasts and human bladder cancer cells, processes that are mediated by Janus kinase/c-Jun inhibition (49). Clinical analysis of invasive human breast cancer revealed a marked correlation between PPARGC1A expression in invasive cancer cells and the formation of distant metastases (50). The silencing of PPARGC1A in cancer cells suspended their invasive potential and attenuated metastasis (50). Although specific data from these previous studies have established that the increase of certain let-7e candidate targets may be promotion signals for cancer metastasis, these genes may also have opposite functions in other contexts.
In summary, it was demonstrated that hsa-let-7e-5p was differentially expressed in primary tumor tissues of RC with heterochrony hepatic metastases (HHM). Furthermore, hsa-let-7e-5p may be used as a prognostic marker to identify patients with RC who may be at risk of metastases.
Acknowledgements
The authors would like to thank Dr Jin Gu (Peking University School of Oncology, Beijing, China) for advice and a critical reading of the manuscript, and the Department of Colorectal Surgery, Peking University School of Oncology, Beijing Institute for Cancer Research, Beijing Cancer Hospital (Beijing, China) for the original sample collection and the use of these samples in the present study.
Funding
The present study was supported by the National Nature Science Foundation of China (grant no. 81172380), the Special Project on the Integration of Industry and Education of Fujian Province (grant no. 2010Y4006), the National Natural Science Foundation of China (grant no. 31601894), the Startup Funds from Fuzhou University (grant no. XRC1463), the Education and Scientific Research Projects of Young Teachers in Fujian Province (grant no. JAT160071), the Fujian Natural Science Foundation (grant. no. 2017J0106).
Availability of data and materials
The datasets generated and analyzed in the present study are included in this published article and are available from the corresponding author upon request.
Authors' contributions
YZ and YuY conceived the project and supervised the experiments. WC, GL, HS, XWa, QY, XWe, LS, FC, and SY performed sample collection, RNA extraction and qPCR. WC, YiY and JC performed cell studies. WC, GL, YuY and YZ performed data analysis and wrote the manuscript, with contributions from the other authors.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of Peking University School of Oncology. Written informed content was obtained from all patients.
Consent for publication
Written informed content was obtained from all patients prior to enrollment in the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Stangl R, Altendorf-Hofmann A, Charnley RM, Scheele J. Factors influencing the natural history of colorectal liver metastases. Lancet. 1994;343:1405–1410. doi: 10.1016/S0140-6736(94)92529-1. [DOI] [PubMed] [Google Scholar]
- 4.Geramizadeh B, Robertson S. Serrated polyps of colon and rectum: A clinicopathologic review. J Gastrointest Cancer. 2017;48:291–298. doi: 10.1007/s12029-017-9977-y. [DOI] [PubMed] [Google Scholar]
- 5.Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang Z, Xi J, Yan L, Gu J. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86. doi: 10.1186/1476-4598-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Van Nhieu Tran J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu T, Weng S, Tang W, Xue R, Chen S, Cai G, Cai Y, Shen X, Zhang S, Dong L. Overexpression of BIRC6 is a predictor of prognosis for colorectal cancer. PLoS One. 2015;10:e0125281. doi: 10.1371/journal.pone.0125281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 10.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 11.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Du L, Wen Z, Yang Y, Li J, Wang L, Zhang X, Liu Y, Dong Z, Li W, et al. Up-regulation of miR-182 expression in colorectal cancer tissues and its prognostic value. Int J Colorectal Dis. 2013;28:697–703. doi: 10.1007/s00384-013-1674-0. [DOI] [PubMed] [Google Scholar]
- 13.Shivapurkar N, Mikhail S, Navarro R, Bai W, Marshall J, Hwang J, Pishvaian M, Wellstein A, He AR. Decrease in blood miR-296 predicts chemotherapy resistance and poor clinical outcome in patients receiving systemic chemotherapy for metastatic colon cancer. Int J Colorectal Dis. 2013;28:887. doi: 10.1007/s00384-012-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T, Zhang G, Liu Z, Xia S, Tian H. Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int J Colorectal Dis. 2013;28:19–24. doi: 10.1007/s00384-012-1528-1. [DOI] [PubMed] [Google Scholar]
- 15.Rotelli MT, Di Lena M, Cavallini A, Lippolis C, Bonfrate L, Chetta N, Portincasa P, Altomare DF. Fecal microRNA profile in patients with colorectal carcinoma before and after curative surgery. Int J Colorectal Dis. 2015;30:891–898. doi: 10.1007/s00384-015-2248-0. [DOI] [PubMed] [Google Scholar]
- 16.Jr CMT, Beauchamp RD, Evers BM, Mattox KL, editors. Sabiston Textbook of Surgery: The Biological Basis of Modern Surgical Practice. 18th edition. Saunders; 2008. [Google Scholar]
- 17.Benson AB, III, Venook AP, Bekaii-Saab T, Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ, Grem JL, et al. Rectal cancer, version 2.2015. J Natl Compr Canc Netw. 2015;13:719–728. doi: 10.6004/jnccn.2015.0087. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YJ, Xie YT, Gu J, Yan L, Guan GX, Liu X. Overexpression of cyclin E isoforms correlates with poor prognosis in rectal cancer. Eur J Surg Oncol. 2011;37:1078–1084. doi: 10.1016/j.ejso.2011.08.139. [DOI] [PubMed] [Google Scholar]
- 19.Edge SB, Compton CC. The american joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 20.Tapan U, Ozbayrak M, Tatli S. MRI in local staging of rectal cancer: An update. Diagn Interv Radiol. 2014;20:390–398. doi: 10.5152/dir.2014.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai X, Liu G, Lu Y, Yin W. Accuracy of endoscopic ultrasound in the preoperative staging and the guidance of transanal endoscopic microsurgery for rectal cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2015;18:487–490. (In Chinese) [PubMed] [Google Scholar]
- 22.Kwak Y, Lee HE, Kim WH, Kim DW, Kang SB, Lee HS. The clinical implication of cancer-associated microvasculature and fibroblast in advanced colorectal cancer patients with synchronous or metachronous metastases. PLoS One. 2014;9:e91811. doi: 10.1371/journal.pone.0091811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engstrom PF, Arnoletti JP, Benson AB, III, Chen YJ, Choti MA, Cooper HS, Covey A, Dilawari RA, Early DS, Enzinger PC, et al. NCCN clinical practice guidelines in oncology: Rectal cancer. J Natl Compr Canc Netw. 2009;7:838–881. doi: 10.6004/jnccn.2009.0057. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs RT, Sun Z, Zhuang F, Robb GB. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS One. 2015;10:e0126049. doi: 10.1371/journal.pone.0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Peng W, Li Z, Li W, Li L, Pan J, Zhang S, Miao Y, Chen S, Su S. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: Comparison between nurses and foragers. Insect Mol Biol. 2012;21:297–303. doi: 10.1111/j.1365-2583.2012.01135.x. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 27.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. (Web Server Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking‘ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Dweep H, Gretz N, Sticht C. miRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Radisky DC, Yang D, Xu R, Radisky ES, Bissell MJ, Bishop JM. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol. 2012;14:567–574. doi: 10.1038/ncb2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavert N, Shvab A, Sheffer M, Ben-Shmuel A, Haase G, Bakos E, Domany E, Ben-Ze'ev A. c-Kit is suppressed in human colon cancer tissue and contributes to L1-mediated metastasis. Cancer Res. 2013;73:5754–5763. doi: 10.1158/0008-5472.CAN-13-0576. [DOI] [PubMed] [Google Scholar]
- 35.Aytes A, Mitrofanova A, Kinkade CW, Lefebvre C, Lei M, Phelan V, LeKaye HC, Koutcher JA, Cardiff RD, Califano A, et al. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc Natl Acad Sci USA. 2013;110:E3506–E3515. doi: 10.1073/pnas.1303558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Pu X, Shi M, Chen L, Song Y, Qian L, Yuan G, Zhang H, Yu M, Hu M, et al. Critical role of c-Jun overexpression in liver metastasis of human breast cancer xenograft model. BMC Cancer. 2007;7:145. doi: 10.1186/1471-2407-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ness TL, Ewing JL, Hogaboam CM, Kunkel SL. CCR4 is a key modulator of innate immune responses. J Immunol. 2006;177:7531–7539. doi: 10.4049/jimmunol.177.11.7531. [DOI] [PubMed] [Google Scholar]
- 38.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang H, Ward WF. PGC-1alpha: A key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 40.Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, Nam SJ, Chun KH. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14:R14. doi: 10.1186/bcr3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang S, He L, Zhao X, Miao Y, Gu Y, Guo C, Xue Z, Dou W, Hu F, Wu K, et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One. 2011;6:e18409. doi: 10.1371/journal.pone.0018409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu X, Guo J, Zheng L, Li C, Zheng TM, Tanyi JL, Liang S, Benedetto C, Mitidieri M, Katsaros D, et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol Cancer Res. 2013;11:240–250. doi: 10.1158/1541-7786.MCR-12-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun J, Frankenberger CA, Kuo WL, Boelens MC, Eves EM, Cheng N, Liang H, Li WH, Ishwaran H, Minn AJ, Rosner MR. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30:4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 46.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 47.Liao DJ, Dickson RB. c-Myc in breast cancer. Endocr Relat Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 48.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 49.Fang Y, Wang Y, Wang Y, Meng Y, Zhu J, Jin H, Li J, Zhang D, Yu Y, Wu XR, Huang C. A new tumour suppression mechanism by p27Kip1: EGFR down-regulation mediated by JNK/c-Jun pathway inhibition. Biochem J. 2014;463:383–392. doi: 10.1042/BJ20140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeBleu VS, O'Connell JT, Herrera Gonzalez KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Chinen Domingos LT, Rocha RM, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(992–1003):1–15. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed in the present study are included in this published article and are available from the corresponding author upon request.