Figure 3.
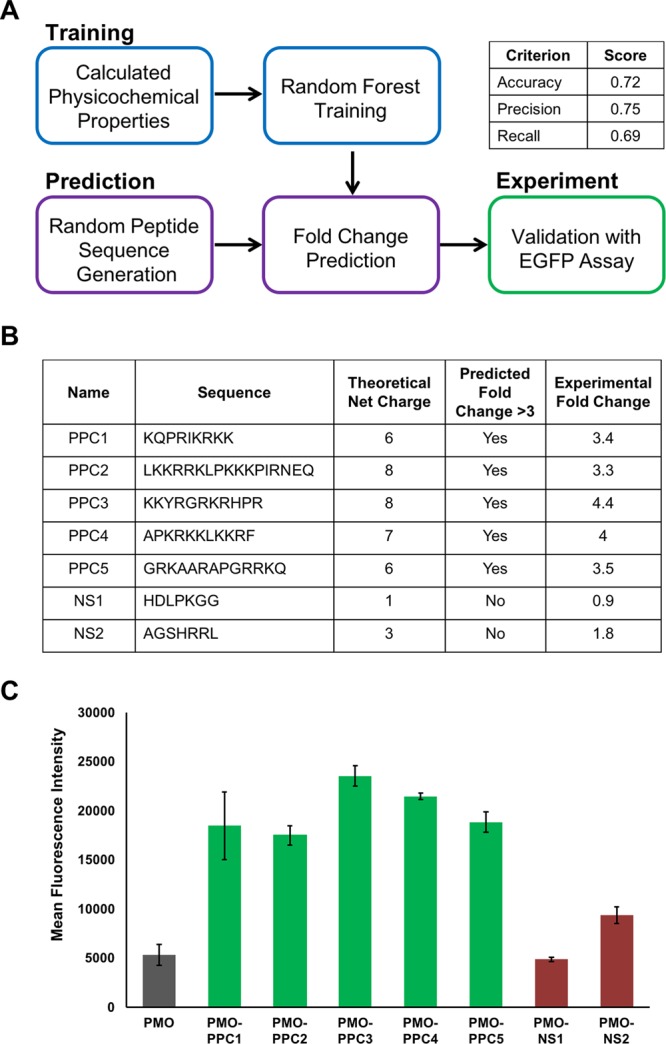
Random forest ensemble learning methods can be used to predict peptide sequences that facilitate PMO delivery. (A) Scheme of the workflow for the development of computationally derived peptide sequences for antisense delivery. One component of the workflow was random forest training using the properties of the CPPs in the library to build a model. Another component was randomly generating peptide sequences and using the model to predict the peptide activity. Lastly, select sequences were synthesized for experimental validation. Performance metrics for the model based on the test set are given in the table. (B) Table of five predicted PMO carriers (PPC) and two predicted negative sequences (NS). All five PMO–PPC conjugates exhibited above a 3-fold improvement in eGFP fluorescence, whereas both PMO–NS conjugates exhibited less than a 3-fold change with respect to PMO. (C) The mean fluorescence intensity of eGFP HeLa cells treated at a concentration of 5 μM with each of the PMO–PPCs and PMO–NSs in serum-containing media. Each individual experiment consisted of the average of three different wells with the same treatment conditions, and the experiment was repeated three times. The error bars represent the standard deviation across the experimental replicates.
