Figure 5. Diabetic wound CD11b+Ly6CHi and CD11b+Ly6CLo monocyte/macrophages display distinct transcriptome profiles.
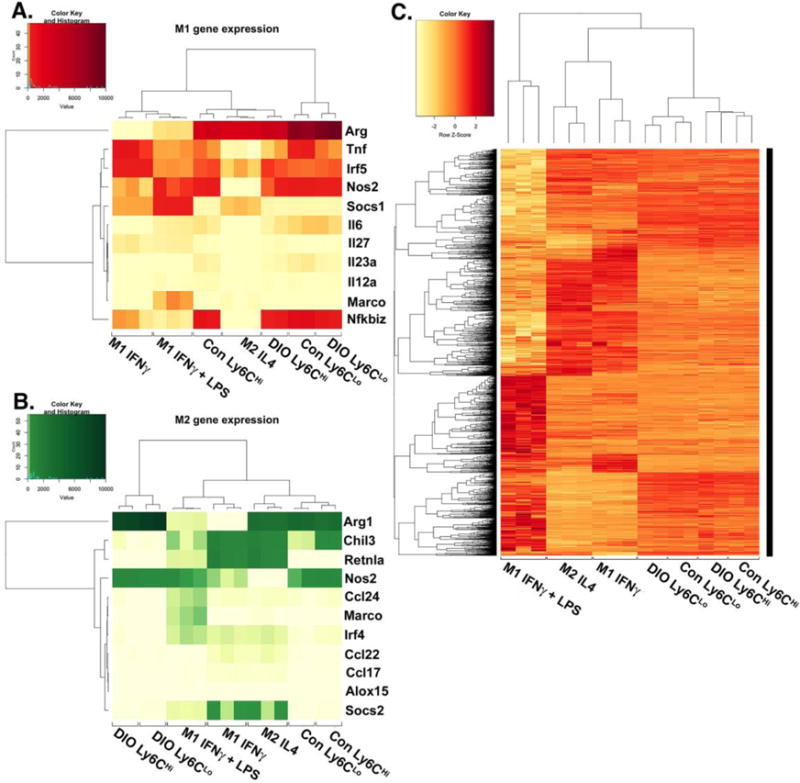
Wounds from normal and high fat diet (HFD)-fed, diet-induced obese (DIO) C57BL/6 mice were collected on post-injury day 3 for cell isolation, surface staining, and FACs to isolate Ly6CHi[Lin−Ly6G−CD11b+] and Ly6CLo[Lin−Ly6G−CD11b+] monocyte/macrophages for RNA-sequencing (RNA-seq). RNA samples were processed by the NIH-funded, University of Michigan DNA sequencing core. Reads were trimmed using Trimmomatic and mapped using HiSAT2. Read counts were performed using the featureCounts option from the subRead package followed by the elimination of low reads and normalization using edgeR. For comparison to in vitro generated macrophages, M1[IFNγ], M1[LPS,IFNγ] and M2[IL-4] datasets were used from publically available transcriptome data generated by Piccolo et al44. (A) A heatmap of normalized reads obtained from edgeR for an internationally recognized panel of consensus genes for in vitro generated M1 macrophages. Wound Ly6CHi[Lin−Ly6G−CD11b+] and Ly6CLo[Lin−Ly6G−CD11b+] monocyte/macrophages were compared to the M1[IFNγ], M1[LPS,IFNγ], and M2[IL-4] transcriptomes. (B) A heatmap of normalized reads obtained from edgeR for an internationally recognized panel of consensus genes for in vitro generated M2 macrophages. Wound Ly6CHi[Lin−Ly6G−CD11b+] and Ly6CLo[Lin−Ly6G−CD11b+] monocyte/macrophages were compared to the M1[IFNγ], M1[LPS,IFNγ], and M2[IL-4] transcriptome. (C) Clustered Pearson correlation of the entire transcriptome of in vivo DIO and control Ly6CHi[Lin−Ly6G−CD11b+] and Ly6CLo[Lin−Ly6G−CD11b+] monocyte/macrophages and in vitro M1[IFNγ], M1[LPS,IFNγ], and M2[IL-4] macrophages (n = 8 mice per group; 4 wounds per mouse were pooled to obtain a single biological replicate given low RNA volumes. Two biological replicates per group were examined.)
