Abstract
Identification of primate hippocampal subfields in vivo using structural MRI imaging relies on variable anatomical guidelines, signal intensity differences, and heuristics to differentiate between regions (Yushkevich et al., 2015a). Thus, a clear anatomically-driven basis for subfield demarcation is lacking. Recent work, however, has begun to develop methods to use ex vivo histology or ex vivo MRI (Adler et al., 2014; Iglesias et al., 2015) that have the potential to inform subfield demarcations of in vivo images. For optimal results, however, ex vivo and in vivo images should ideally be matched within the same healthy brains, with the goal to develop a neuroanatomically-driven basis for in vivo structural MRI images. Here, we address this issue in young and aging rhesus macaques (young n=5 and old n=5) using ex vivo Nissl-stained sections in which we identified the dentate gyrus, CA3, CA2, CA1, subiculum, presubiculum, and parasubiculum guided by morphological cell properties (30 μm thick sections spaced at 240μm intervals and imaged at 161 nm/pixel). The histologically identified boundaries were merged with in vivo structural MRIs (0.625 × 0.625 × 1 mm) from the same subjects via iterative rigid and diffeomorphic registration resulting in probabilistic atlases of young and old rhesus macaques. Our results indicate stability in hippocampal subfield volumes over an age range of 13 to 32 years, consistent with previous results showing preserved whole hippocampal volume in aged macaques (Shamy et al., 2006). Together, our methods provide a novel approach for identifying hippocampal subfields in non-human primates and a potential “ground truth” for more accurate identification of hippocampal subfield boundaries on in vivo MRIs. This could, in turn, have applications in humans where accurately identifying hippocampal subfields in vivo is a critical research goal.
Introduction
An important goal for behavioral and systems neuroscientists is to translate knowledge gained from nonhuman animals into a deeper understanding of the human brain and cognitive processes. Leyla de Toledo-Morrell leaves behind a solid legacy of scientific discovery and achievement in this field through her work on animal models of normative brain aging, as well as on normal and pathological human aging. She published some of the earliest electrophysiological and cognitive work on the aging rat (de Toledo-Morrell and Morrell, 1985) and then moved her focus to human aging and imaging methodologies. Her work illustrates how she was conceptually inspired by results from basic science experiments in rodents – and then used these results to generate and test hypotheses about normal human brain aging (Rogalski et al., 2012; Stoub et al., 2012). These experiments provide strong support for the idea that axons from the entorhinal cortex projecting to the hippocampus are pruned as part of the normative biological aging process in mammals. Leyla de Toledo-Morrell also made important research contributions to the field of Alzheimer’s disease. In a recent study she and her colleagues identified methods to detect very early signs of disease onset that give great hope for testing and detecting brain changes in response to early treatment interventions (Stoub et al., 2014). In this vein, we believe the data presented here reflect questions that would have captured her scientific curiosity, and of course are in the spirit of her commitment to translating data collected in animal models to inform analytic approaches to brain structure and function in humans.
There have been a number of large recent efforts directed towards more accurately identifying boundaries that define the entire hippocampus (Frisoni and Jack, 2015) or even the subfields within the structure (Adler et al., 2014; Berron et al., 2017; Iglesias et al., 2015; Wisse et al., 2016a; Yushkevich et al., 2015a; Yushkevich et al., 2015b). This is a particularly important effort for a number of reasons, including the fact that there is substantial evidence that the component parts of the hippocampus and surrounding regions of cortex make unique computational contributions to the circuit. In particular, evidence from human neuroimaging suggests that hippocampal subfields play distinct roles in memory processing, particularly pattern separation and completion (Bakker et al., 2008; Stokes et al., 2015), converging with multiple rodent studies (Guzowski et al., 2004; Neunuebel and Knierim, 2014). Furthermore, there appear to be differences in which hippocampal subfields are most susceptible to age-related neural decline in humans (Vieweg et al., 2015; Yassa et al., 2011), monkeys (Thome et al., 2016), and rats (Wilson et al., 2005). Thus, individual deterioration of these components in aging, or in age-related diseases, would be expected to have different cognitive consequences. Therefore, obtaining a better understanding of hippocampal subfield anatomy and physiology, and how it might differ as a function of aging, is an important research goal on several fronts.
In humans, numerous attempts have been made to define the boundaries of hippocampal subfields in vivo (Berron et al., 2017; Ekstrom et al., 2009; Kerchner et al., 2012; Mueller et al., 2007; Suthana et al., 2015; Yushkevich et al., 2015a; Zeineh et al., 2000; Zeineh et al., 2001). A recent paper, however, illustrated significant differences in the agreement and alignment of different high-resolution hippocampal MRI subfield tracing procedures (Yushkevich et al., 2015a), potentially challenging some of these efforts. While the findings from Yushkevich et al. indicate inconsistency of subfield labeling protocols used by different laboratories to demarcate regions using structural MRI, these differences could come about for a variety of reasons.
One factor that could account for the discrepancies seen in Yushkevich et al. is inter-rater reliability, in which different individuals demarcate subfields at different places. Most inter-rater reliability scores within a laboratory, however, are typically high (i.e., dice coefficients of around 0.75–0.95 (Berron et al., 2017; Yushkevich et al., 2015a)). Therefore, a difference in the protocols themselves, is the more likely explanation for the differences observed between laboratories (Yushkevich et al., 2015a). A likely contributor to the differences in protocols between labs is a lack of clear signal/contrast intensity cues in the MRI capable of indicating boundaries between subfields. In support of this view, recent work utilizing human in vivo and ex vivo MRI, and histology for two subjects, illustrated that the popular automated segmentation tool, ASHS, makes systematic errors when drawing the boundary between CA1 and Subiculum (Wisse et al., 2016a). Because the ASHS algorithm relies on a collection of hand labeled MRIs, this bias is likely attributable to an overreliance on geometric rules rather than anatomical cues in the protocol used to trace the algorithm’s “multi-atlas” bank of example data.
An additional complicating factor in accurately labeling hippocampal subfields in vivo are the differences in subfield locations between subjects. Without clear signal/contrast intensity cues in the MRI, the vast majority of subfield protocols are, instead, based on histologically identified subfield locations in single subjects. The anatomy of these single subjects are typically summarized with geometric rules (Entis et al., 2012; Mueller et al., 2007; Pluta et al., 2012) which are then applied to label in vivo MRIs from other individuals. Thus, the need for anatomically-driven guidelines are critical issues to address, and in fact, are the focus of ongoing efforts of the Hippocampal Subfield Segmentation Summit group, which seeks to develop guidelines for identifying human hippocampal subfields (Wisse et al., 2016b).
Recent work has begun the task of moving toward consistent, anatomically-based labels that capture individual variability by merging human ex vivo MR imaging and histology with in vivo MRI images (Goubran et al., 2015; Wisse et al., 2016a), providing key insight into accurately identifying hippocampal subregions in vivo. Two key issues, however, remain as significant challenges for this work. One challenge is that the human hippocampus shows considerable variability between subjects that complicates the task of correctly labeling tissue and generalizing across individuals. For instance, distinct patterns of folding in perirhinal cortex and differences in the number of digitations in the hippocampal head have made accurate identification of subfields a challenge (Ding and Van Hoesen, 2010; Ding and Van Hoesen, 2015). Second, although some efforts have involved in vivo MRIs and ex vivo histological sections in the same human subject, these efforts are rare and have been limited to patient populations prone to selective hippocampal atrophy (Goubran et al., 2015; Wisse et al., 2016a). Due to these constraints, considerable challenges remain for acquiring, labeling and processing datasets capable of fully capturing individual variability, describing hippocampal anatomy at all ages and sexes and accounting for patient-specific pathology.
Traditionally, the complications associated with studying humans have been partially alleviated by turning to nonhuman primate animal models. The anatomy of the rhesus macaque brain is better understood compared with humans (Amaral and Lavenex, 2007), and although studies comparing human and macaque anatomy are far from comprehensive, the existing data suggest overwhelming similarity in relative cell density, cytoarchitecture, chemoarchitecture, and neural connectivity of the two species (Amaral and Lavenex, 2007; Amaral et al., 2007; Ding, 2013; Ding et al., 2010; Lim et al., 1997). Additionally, healthy subjects both young and old can easily be compared, shedding light on whether anatomical changes occur in healthy aging. While there are neuroanatomical differences between monkey and human hippocampus, these are generally regarded as fairly subtle compared to other species like rats (Amaral and Lavenex, 2007; van Dijk et al., 2016). Thus, developing protocols for matching histology to MRI in young and aged macaque brains is an important step not only towards a better understanding the monkey hippocampus, but has the potential to complement and inform similar efforts in humans because they involve healthy brains.
Here we describe a novel approach to demarcate subfields in the rhesus macaque based on Nissl-stained sections. These labels are then projected onto in vivo MRIs from the same monkey by registering the histological images to the structural scans. Together, this approach offers a new method for labeling hippocampal subfields of nonhuman primates on MRIs based on histological definitions, with the ability to compare between age groups and potentially to extrapolate to humans.
METHODS
Subjects
Five young adult (13.4 – 17.6 years) and five aged (24.6 – 31.7 years) rhesus monkeys (Macaca mulatta) served as subjects. Groups were made up of both females (2 young and 1 aged) and males (3 young and 4 aged). The age of each monkey can be multiplied by 3 to provide an approximate comparison to equivalent human ages (Tigges et al., 1988). All of the animals were housed in pairs and were provided environmental enrichment including monitored socialization in a large outdoor pen, behavioral testing and regular fruits and vegetables.
Laboratory primate chow was provided on a twice-daily regimen without dietary restriction. Water was provided in the home cage on an ad libitum basis and the primate colony was kept on an alternating 12 h light/dark cycle schedule. All experimental procedures were performed in accordance with the National Institutes of Health Guidelines and were approved by Institutional Animal Care and Use Committees at the California National Primate Research Center and the University of California, Davis.
Magnetic resonance imaging
Monkeys were anesthetized with ketamine (20 mg/kg, intra muscular (i.m.)), dexametatomidine (30 mcg/kg i.m.) and atropine (0.004 mg/kg i.m.). The subject’s head was then placed in an MRI compatible stereotaxic frame and positioned within the standard quadrature radiofrequency coil used for human imaging. Eighty contiguous 1 mm coronal T1-weighted images of the whole brain were acquired for each monkey on a 1.5 T GE Sigma Horizon L NV/I MRI system (GE Medical Systems) with version 84 M4 software at the UC Davis imaging Research Center, Sacramento, CA. A radio frequency spoiled gradient recalled echo sequence (3D SPGR) was used with the following parameters: time repetition 21 ms, time echo 7.9 ms (full echo), flip angle 30°, field of view 16 × 16 cm, acquisition matrix 256 × 256, NEX 4 (no phase wrap option), and bandwidths 1563kHz for a resolution of 0.625 × 0.625 × 1 mm. Each monkey’s anesthesia was reversed with atipamezole (30 mcg/kg) post scan. The gradient subsystem of the MRI system was measured prior to and throughout the study period. Local GE service personnel calibrated the subsystem monthly. Geometric accuracy was documented as stable to within ±0.15% (i.e., ± 0.15 mm per 10 cm). Adjustments were not necessary to maintain that level of accuracy and precision throughout the study period.
Histological procedures
At necropsy, the subjects were transcardially perfused for 2 min with 1% paraformaldehyde in 0.1 M phosphate buffer, followed by an 8 min perfusion with 4% paraformaldehyde in 0.1 M phosphate buffer, and finally a 50 min perfusion with 4% paraformaldehyde in 0.1 M phosphate buffer. After removal from the skull, the brains were post-fixed in 4% paraformaldehyde in 0.1 M phosphate buffer. Fixation time spanned between 10 and 367 days (see Supplemental Table 1 for complete data) and did not significantly differ between old and young subjects. Brains were then blocked in the coronal plane caudal to the retrosplenial cortex. The two blocks of each brain were cryoprotected prior to freezing by placing them into 10% glycerol with 2% DMSA in 0.1M phosphate buffer for 24 hours and then into 20% glycerol with 2% DMSO in 0.1M phosphate buffer for 72 hours. The brains were frozen in isopentane chilled with dry ice in an ethanol bath. The brains were coronally sectioned at 30 μm on a sliding microtome. Sections were collected in four series, one of which was Nissl-stained for the entire extent of the brain. Nissl-stained slides through the hippocampal region were scanned (each monkey contributing between 67-83 sections) using an Olympus VS 110 Virtual Slide Microscope system at the California National Primate Research Center. Scans were performed with an Olympus PlanSApo 10×/0.40 lens and an Olympus XC10 camera. These scanned sections were then used for subregion tracing to cover hippocampal regions corresponding to the area of the brain between plates 60-98 in the (Paxinos G, 2009) atlas of the rhesus macaque.
Subregion boundary identification
We employed guidance from the following anatomical references to define the different subcomponents comprising the monkey hippocampus (Amaral and Lavenex, 2007; Buckmaster and Amaral, 2001). Figure 1 provides a step-by-step set of procedures for visual demarcation of the hippocampal subfields (also see Supplemental Figure 1). Below, we also provide a text-based description that walks the reader through the visual-based description.
Figure 1. Subfield labeling protocol.
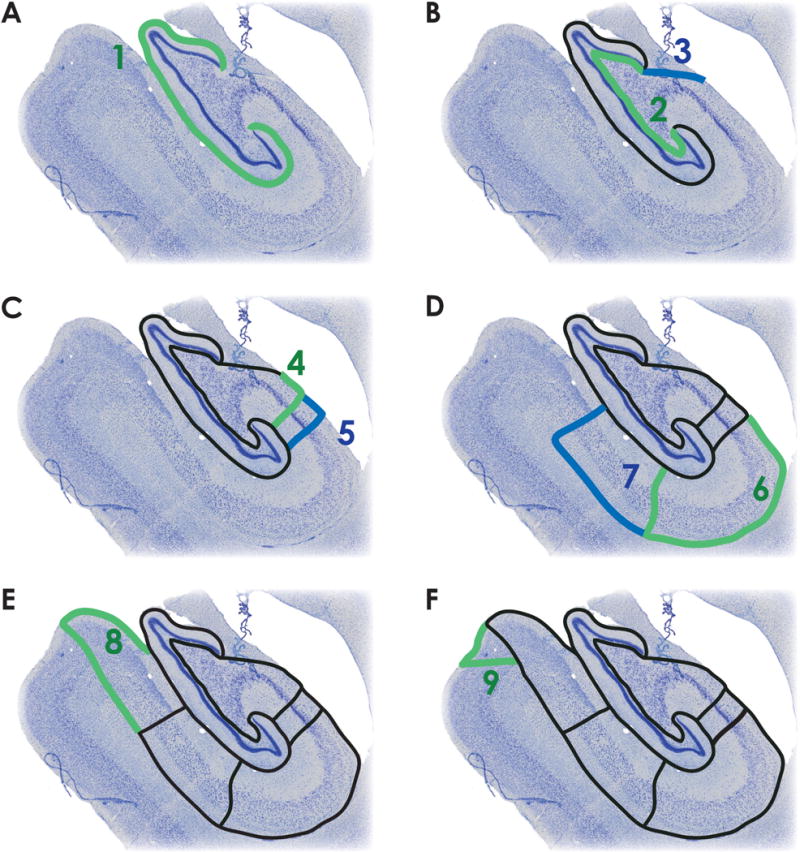
A. The outer boundary of the DG (dentate gyrus) is defined by the hippocampal fissure (green, step 1). B. The thin polymorphic layer, which lies adjacent to the granule cell layer (green, step 2), defines the inner boundary of DG. The fimbria is delineated from CA3 by connecting the medial blade of DG with alveus/lateral ventricle border (blue, step 3). C. The CA3/CA2 border is marked by the dense pyramidal cell layer of CA2, and is drawn parallel to the pyramidal cells of CA2/CA3 (green, step 4). The CA2/CA1 border is marked by a transition to the diffuse pyramidal cell layer of CA1 (blue, step 5). D. CA1 is defined by the alveus along the temporal horn of the lateral ventricle and along the alveus/ILF (inferior longitudinal fasciculus) border to the CA1/subiculum boundary, marked by a reduction in the presence of bipolar neurons and a change in the density and orientation of pyramidal cells (green, step 6). Subiculum is defined by the alveus/ILF border until the alveus boundary is lost. After the alveus, it’s approximate thickness is preserved relative to the cell layer. The subiculum/presubiculum boundary marked by the densely packed cells of the presubiculum (blue, step 7). E. The presubiculum is labeled by continuing along the ILF to the presubiculum/parasubiculum boundary, marked by a change in cell density (green, step 8). F. The parasubiculum/parahippocampal cortex boundary is marked by the transition to layered cortex. The cells of the parasubiculum are bound medially by laminar cortex and laterally by presubiculum (green, step 9).
We began with a mid-level rostral-caudal section through the hippocampus (Figure 1) as follows:
Draw the outer boundary of the dentate gyrus (DG). Draw a line along the edge of the hippocampal fissure, which shares a border with the mostly cell free molecular layer of DG. Extend the line along the sulcus until reaching the granule cell layer in the upper and lower blades of the DG (Figure 1A).
Identify the DG/CA3 boundary by continuing the prior boundary line along the inner edge of the DG blades, following along the granule cell layer. Included in this region is the thin polymorphic layer running next to the granule cell layer, which is characterized by “football-shaped” bipolar cells. This boundary separates DG from the adjacent CA3 subfield (Figure 1B, green line).
Identify the fimbria/CA3 boundary by separating the fimbria from the CA3 field. Do this by drawing a line from the end of the medial blade of the granule cells to the alveus/lateral ventricle border (Figure 1B, blue).
Identify the CA3/CA2 boundary. The CA2 field is characterized by a denser pyramidal cell layer due to the characteristic multiple compact stacks of cells. Continue to draw a line along the alveus/lateral ventrical border. Once the CA3/2 boundary is reached, draw a line perpendicular to the hippocampal fissure (Figure 1C, green).
Identify the CA2/CA1 boundary based on the transition from the dense pyramidal cell layer of the CA2 field to the diffuse pyramidal cell layer of CA1. In a similar fashion to the CA3/CA2 boundary, draw a line perpendicular to the hippocampal fissure (Figure 1C, blue).
Identify the CA1/subiculum boundary by extending the CA1/ventricle border line along the alveus to the boundary of CA1/subiculum where there is a reduction in the presence bipolar interneurons. Identify the CA1/subiculum boundary based on a change in the density and direction of the pyramidal cell dendrites. Extend the line along the boundary to the hippocampal fissure (Figure 1D, green).
Identify the subiculum/presubiculum boundary by locating the densely packed cells of the presubiculum. Continue to draw a line beginning at the juncture of the alveus and CA1 border. Continue the line along the axis of the alveus through the white matter to the presubiculum, which is characterized by a compact group of small cells arranged in a triangular shape. At the lateral tip of the triangle, draw a line perpendicular to the hippocampal fissure and the CA1/subiculum boundary (Figure 1D, blue).
Identify the presubiculum/parasubiculum boundary by locating the region where the cellular density changes between presubiculum and parasubiculum. Continue drawing the line at the junction of the subiculum/presubiculum border by extending it to the surface of the brain and looping back to the hippocampal fissure (Figure 1E, green).
Identify the parasubiculum/parahippocampal cortex boundary by drawing a line at the surface of the brain and ending in the transition to layered cortex. A line is then drawn encompassing the cells of the parasubiculum, which are bound medially by laminar cortex and laterally by presubiculum (Figure 1F, green).
We refer the reader to Supplemental Figure 1, where we have included a standalone document that depicts in more detail the subfield boundaries. Subfield drawing was performed in Adobe Photoshop. After loading a histological section, a new layer was added to the image and the boundaries of each subfield were marked according to the protocol presented above on the new layer. After tracing, each subfield was filled in using a unique color to mark each of the subfields (Figure 2). After a section was completed, a Photoshop script saved the tracing layer as a new image. Further processing converted each subfield into a separate binary mask and down-sampled these subfield demarcations along with the histology for further processing (see down-sampling histological images for resolution details). Importantly, subfield label images followed the same transformations applied to the histological images for all steps in the analysis pipeline.
Figure 2. Example subfield boundaries from one individual monkey.
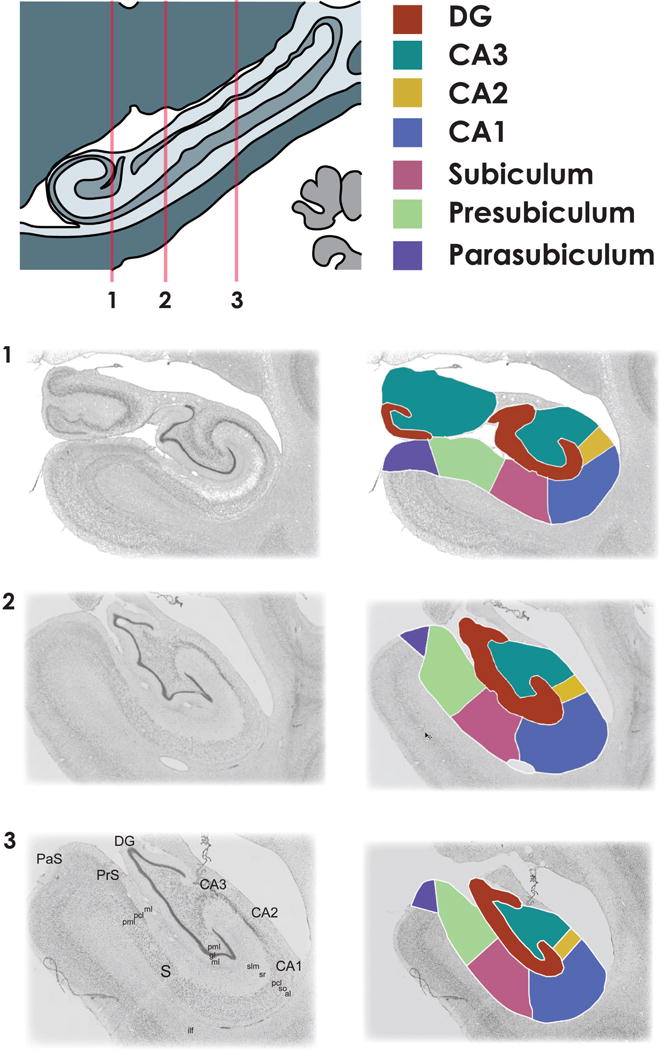
Subfield boundaries drawn at three anterior-posterior locations along the extent of the hippocampus (1-most anterior, 3-most posterior). Once a subject’s boundaries are completed, each subfield is assigned a unique identifying color. Multicolor mask images are then converted to separate binary images for each hippocampal subfield for further processing.
[pml - polymorphic layer, gl - granule cell layer, slm - stratum lacunosum-moleculare, sr – stratum radiatum, pcl - pyramidal cell layer, so - stratum oriens, al – alveus, ilf - inferior longitudinal fasciculus]
Subfield demarcation inter-rater reliability assessment
To establish the reliability of our demarcation protocol, 3 raters segmented each subfield on one histologically sectioned monkey hippocampus. Inter-rater reliability was measured for each subfield using the Sorenson-Dice similarity coefficient. This metric is defined as 2 times the overlapping-area between two raters divided by the sum of each rater’s total area, as below:
Coefficients for each pair of raters were computed for each section and each subfield using the full-resolution binary images described above. The average Sorenson-Dice metric across rater-pairs served as the basis of our inter-rater reliability score.
Histological preprocessing steps
In order to attain our goal of aligning the histology to MRI images, a series of preprocessing and image registration steps were performed. These steps involved down-sampling histology images to computationally tractable resolutions, performing 2D rigid registration to align sections into an orderly stack, non-rigid 2D registration to correct for inconsistencies in the tissue mounting process, converting the histology image stack to a 3D image, and finally registering the histology to the MRI via diffeomorphic registration.
Down-sampling histological images
Nissl-stained sections were scanned at a resolution of 644.16 by 644.16 nm and .24mm between sections. To prepare for subfield demarcation and image processing each slide image was converted from Olympus’ proprietary “vsi” format to a tiff format using the bio-formats “loci_tools.jar” java library (http://www.loci.wisc.edu/bio-formats/). In order to make disk space requirements more manageable, each image was down-sampled to 2.5766 by 2.5766 μm during the conversion process. Subfield demarcation was conducted at 2.5766 by 2.5766 μm, however this resolution was too high to make the remaining image processing steps tractable. Thus, images were further down-sampled to 51.532 by 51.532 μm and converted to grayscale (via 0.2989*R + 0.5870*G + 0.1140*B) for the remaining image processing steps.
Rigid-registration stack alignment
The first in the series of registration steps was a rigid registration performed between neighboring sections in order to build a contiguous 3D image stack (Figure 3A). The methodology of Adler et al. (2014) was adopted whereby each section is registered to neighbors up to 4 sections away in the stack and graph theory is used to calculate an optimum least-cost set of transformations that serve to register each slide to a central “anchor” section (Figure 3B). As noted by Adler et al., this approach is preferable to simply registering each slide to a neighbor because errors tend to accumulate along the z-direction resulting in drift. This drift can be accentuated by poorly mounted sections, which can cause registration errors and large discontinuities of the image stack if a purely sequential approach is taken. Calculating a least cost path on a graph allows the algorithm to skip sections that were poorly registered resulting in a more continuous stack. Briefly, a chain-like network of all sections is created, with each section serving as a node (Figure 3C, black). Edges are drawn between sections that were registered to one another and the similarity metric acquired during rigid registration is assigned as the edge weight. Finally, Dijkstra’s algorithm finds the least-cost transformation sequence between each slide and the anchor slide (Figure 3C green).
Figure 3. Registration methods.
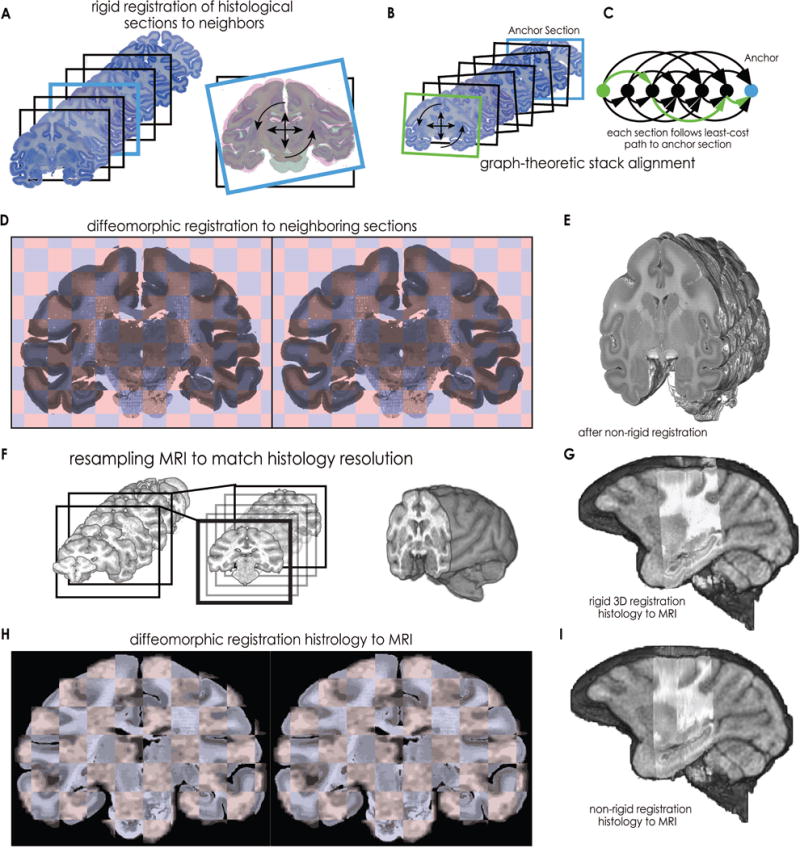
First, histological sections were registered to each other to remove inconsistencies that occurred during mounting (A-D). A. Rigid registrations were computed between each section and neighbors up to 4 sections away in the image stack. For each registration, a measure of similarity between sections was saved for subsequent processing. B. Stack alignment was conducted using the methodology of Adler et al. (2014). Each section (e.g., green) was aligned to a centrally located anchor section (blue) via graph registration. C. A graph was created that reflected the structure of between-neighbor registrations. Each section served as a node, and previously saved similarity values from between-neighbor registrations served as edge weights between nodes (black arrows). Dijkstra’s algorithm then finds the least-cost series of rigid transformations (green arrows) to the anchor section. D. Next, non-rigid diffeomorphic registration between neighboring sections corrects for non-rigid errors in the mounting process, resulting in a contiguous 3D image stack (E). Then the histology was registered to the MRI (F-I). F. To prepare for registration between histology and MRI, lower resolution MRI is up-sampled, first In plane (black box), and then between planes (gray boxes), to better match the high-resolution histology. G. Histology undergoes 9-DOF affine registration to MRI. H. Histology undergoes non-rigid diffeomorphic registration to MRI in 2-D. I. Finally, 3D non-rigid diffeomorphic registration completes the registration process.
Non-rigid tissue warping correction
At this stage in the analysis pipeline, the image stacks were reasonably well aligned into 3D volumes. However, significant discontinuities in the 3D volumes remained due to non-rigid errors in the tissue mounting process such as shearing, shrinking, or folding. To correct for the remaining errors that resisted correction by the rigid graph-theoretic transformation procedure, additional rigid and non-rigid registrations were performed between neighboring sections. Non-rigid registration was performed using the diffeomorphic demons algorithm (Vercauteren et al., 2009). For each section, rigid and diffeomorphic warping estimates were calculated between the section and its immediate neighbors. These warping parameters from each neighbor were averaged and each section was updated from the average warp field (Figure 3D). Rather than sequentially updating each section, which had a tendency to produce compounding registration errors, this procedure was performed on odd sections first, then even sections. This warping procedure resulted in 3D image stacks that were continuous; however, one type of systematic error remained. The introduction of the brainstem in posterior sections caused these sections to become systematically shifted in the dorsal/superior direction. First, we marked the superior-most part of the tissue. Next, we compared these locations to the mean location, which indicated the deviation from a level boundary. Then each section was corrected according to its deviation from the mean. Specifically, the superior-most boundaries were entered into a vector. Next, this vector was demeaned and low pass filtered using a 2.5mm moving average. The values of the filtered boundary location vector were used to shift each section resulting in a 3D tissue stack with a nearly level superior boundary (Figure 3E). The completed image stack was then converted to nifty format using the IO functions included in SPM 12.
MRI preprocessing
First, non-brain tissue was manually removed from each subject’s MRI using fslview. Next, the MRI images were up-sampled from 0.625mm ×.625mm × 1mm to 0.156 mm × 0.156 mm × 0.25 mm (Figure 3F). Because the remaining analysis involved a down-sampling of the histological volumes and subfield tracings to match the resolution of the MRI, up-sampling the MRI aimed to lessen the information loss.
Histology to MRI 3D rigid and non-rigid alignment
In order to align each individual 3D histological volume with its matching MRI volume, a series of preprocessing and registration steps were taken. We first performed manual, affine transformation of the histological volume to the MRI using Slicer 4’s “transforms” editor (Fedorov et al., 2012) (Figure 3G).
To prepare for non-rigid alignment, the contrast and image intensity values between the MRI and histology were matched using an adaptive histogram matching optimization. We calculated a loss function by taking the mean difference in intensity between a middle hippocampal section of MRI and histology. The loss function was minimized by varying the contrast limit parameter in the adaptive histogram equalization procedure for both the MRI and histological section. Optimization was performed using grid search.
After optimizing overall intensity differences between modalities, an additional intensity-matching step was performed to account for the dark appearance of the hippocampus in our T1-weighted MRI images. In this step, mean intensity differences again served as a loss function and grid-search was used to optimize two parameters, kernel size and a multiplier. In order to selectively darken the hippocampal region of the histological images the subfield labels from the dentate gyrus and CA3 regions were combined and assigned a value of 1. Next, these areas were Gaussian smoothed at different kernel sizes, then smoothed images were scaled by a parameter that was multiplied with the smoothed regions. The smoothed regions were subtracted from the histological image, then the loss function was calculated. Grid search optimized both the size of the Gaussian smoothing kernel and the multiplying parameter that optimized the intensity matching between the histology and MRI.
Finally, non-rigid alignment between the MRI and histology was performed using two diffeomorphic warping steps. The first was done in 3D with a relatively large 6mm Gaussian smoothing kernel to provide an initial fit. Then, additional alignment was performed in 2D, with diffeomorphic warping which used a 1mm kernel to register the MRI to the histology in finer detail (Figure 3 H,I, see Supplemental Figure 2 for additional examples).
Group Template Construction
A group template of all individual MRI volumes was constructed using ANTs parallel template construction (Avants et al., 2011). This step computed warping parameters that involved the minimum deformations necessary to non-rigidly register each individual’s MRI to a group average template. The resulting warping parameters were applied to each subject’s subfield labels for further analysis.
Subfield boundary location deviation
For each coronal-plane in the MRIs, boundaries between CA3-CA2, CA2-CA1, CA1-SUB, SUB-preSUB, and preSUB-paraSUB were marked with a single point. The procedure used to calculate each boundary point was to expand two neighboring subfields by 0.5 mm. This resulted in overlap between two regions. The centroid of the area of overlap between two regions was taken to be the boundary. Measuring the degree of deviation around the group mean position for each boundary involved three steps. First, the average boundary position across subjects was calculated for each section. Next, the distance between the average boundary position and each subject’s boundary positions were calculated, giving the deviation at each section. Finally, these deviations were averaged across sections, giving an average absolute deviation for each subject and subfield.
Volume
Subfield volumes were measured in both individual and in group-template space. To correct for overall differences in brain size, subfield volumes were divided by total intracranial volume (ICV) calculated from the skull stripped MRIs. In group-template space volume analysis, a single template for both young and aged subjects was used in order to correct for total brain volume differences between subjects during the registration process. Correcting for individual differences in brain size, either using ICV or group registered brains was necessary before comparing volume between old and young because our age groups did not have the same number of female brains (2 young and 1 aged), which have been shown to have smaller brain volume in the rhesus macaque (Franklin et al., 2000).
RESULTS
Inter-rater reliability of hippocampal subfield labels
To measure the ability of different raters to consistently demarcate the dentate gyrus (DG), CA3, CA2, CA1, subiculum (S), presubiculum (PrS), and parasubiculum (PaS) on histological sections, three raters performed complete demarcations of a single subject’s hippocampal sections. Accuracy was assessed by calculating Sørensen–Dice similarity, defined as the fraction of overlapping pixels divided by the average total number of pixels assigned to a region. Sørensen–Dice similarity was compared between each pair of raters on each hippocampal section for each subfield (Figure 4). Average Sorenson-Dice ranged between 0.92 and 0.74 (DG: 0.91 ± 0.02, CA3: 0.89 ± 0.04, CA2: 0.80 ± 0.07, CA1: 0.92 ± 0.03, S: 0.82 ± 0.05, PrS: 0.87 ± 0.04, and PaS: 0.74 ± 0.13), suggesting similar inter-rater agreement to that seen in MRI based labeling protocols (Berron et al., 2017). Overall, inter-rater reliability was lower for smaller regions such as CA2 and parasubiculum (a common occurrence when using Sørensen–Dice similarity), and was slightly reduced for all regions of the subiculum. This was predicted by our raters, as segmenting these regions was more difficult due to the boundary of the alveus becoming unclear in subicular regions. We return to this issue in the Discussion.
Figure 4. Inter-rater reliability.
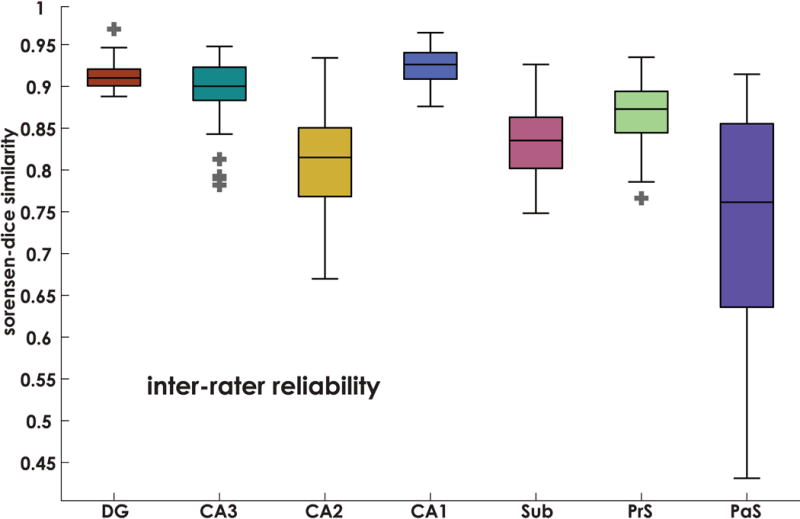
Inter-rater reliability was determined using the Sørensen–Dice similarity method. Here we measured agreement between 3 raters on a single brain. Boxplots denote Sørensen–Dice values for each section of the hippocampus. Average Sorenson-Dice ranged between 0.92 and 0.74 (DG: 0.91 ± 0.02, CA3: 0.89 ± 0.04, CA2: 0.80 ± 0.07, CA1: 0.92 ± 0.03, S: 0.82 ± 0.05, PrS: 0.87 ± 0.04, and PaS: 0.74 ± 0.13), indicating reliability comparable to other tracing protocols (e.g., Berron et al., 2017). Horizontal line in each box indicates the median, boxes denote 25-75% interquartile ranges, whiskers denote 1.5(IQR) boundaries, outliers marked with +.
Absolute deviation of subfield boundaries
To assess the degree of variability in subfield boundary locations in the final template, average absolute deviations were calculated for the boundary locations of neighboring subfields (see Subfield boundary location deviation section). Results are presented in Figure 5. First, to determine whether subfield boundary deviations were significantly different between young and old monkeys, we performed an age group by region repeated-measures ANOVA. Results revealed a main effect of region (F(4,32) = 10.02, p = 2×10−5), but no main effect of group (p = 0.85), or interaction effect (p = 0.09). Post-hoc paired t-tests revealed that the CA3-CA2 boundary deviations were significantly smaller than those of CA2-CA1 (t(9) = 7.37, p = 4.23×10−4, Bonferroni corrected). Absolute deviations spanned between 0.84 and 1.21 mm (CA3-CA2: 0.84 ± 0.28, CA2-CA1: 1.21 ± 0.18, CA1-SUB: 1.14 ± 0.28, SUB-preSUB: 1.07 ± 0.27, preSUB-paraSUB: 1.08 ± 0.23 mm).
Figure 5. Subfield boundary between-subject spatial deviations.
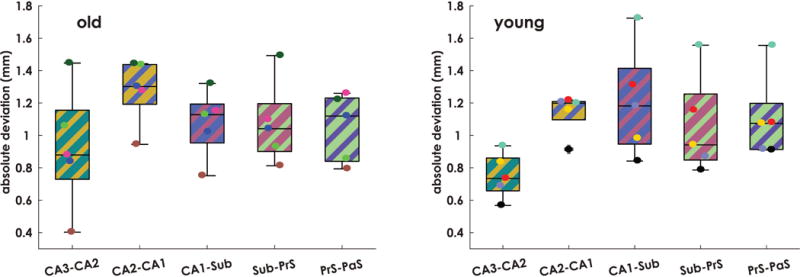
The absolute deviation in subfield boundary locations was quantified for all subjects. Absolute deviations spanned between 0.84 and 1.21 mm (CA3-CA2: 0.84 ± 0.28, CA2-CA1: 1.21 ± 0.18, CA1-SUB: 1.14 ± 0.28, S-PrS: 1.07 ± 0.27, PrS-PaS: 1.08 ± 0.23 mm). An age group by subfield boundary deviation ANOVA revealed a main effect of region (F(4,32) = 10.02, p = 2×10 5), but no main effect of group (p = 0.85), or interaction effect (p = 0.09). Post-hoc paired t-tests revealed that the CA3-CA2 boundary deviations were significantly smaller than those of CA2-CA1 (t(9) = 7.37, p = 4.23×10-4, Bonferroni corrected). The horizontal line in each box indicates the median, boxes denote 25-75% interquartile ranges, whiskers denote 1.5(IQR) boundaries, outliers marked with +.
The resolution of our structural MRI was 0.625 mm in plane, indicating that average absolute deviation fell within 1-2 voxels from center for all subfield boundaries.
Subfield volumes
To address age related change in hippocampal subfield volume, we next calculated total subfield volume for each subject in both individual and group space (see Supplemental Table 1 for raw data). ICV normalized individual-space volumes are shown in Figure 6. Comparing between age groups, Mann-Whitney U-tests indicated that volumes were not different between old and young (p>.67 corrected). Performing the same analysis in group space (Supplemental Figure 3) again revealed no significant differences in subfield volumes between age groups (p>.39, corrected) suggesting that, like whole hippocampal volumetric analysis (Shamy et al., 2006), hippocampal subfield volume is also preserved in aging macaques.
Figure 6. Subfield volume comparison between aged and young rhesus macaques.
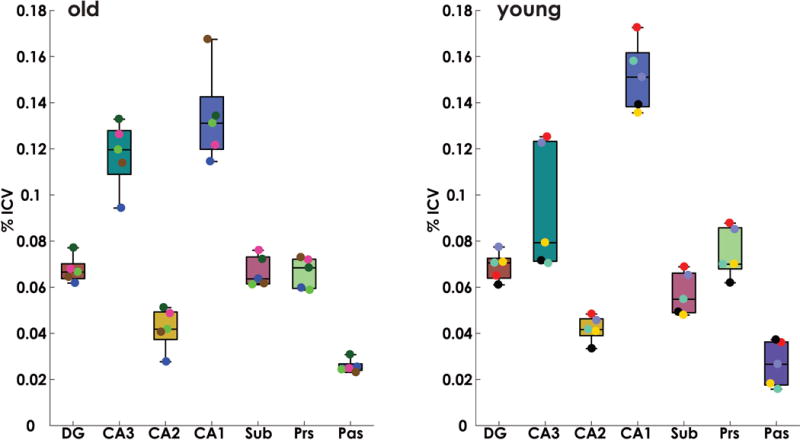
Intracranial volume (ICV) corrected subfield volumes for young and aged animals. Mann-Whitney U-tests were conducted between age groups for each subfield. Results indicated no significant differences in subfield volumes between age groups (p>.67, corrected).
DISCUSSION
Our paper provides several novel contributions helpful for understanding the primate hippocampus. First, we demonstrate the feasibility of reliably labeling primate hippocampal subfields on histological sections. There is currently no agreed upon procedure for labeling nonhuman primate hippocampal subfields and thus our method is an important first step in developing such protocols. Second, we present a novel method for aligning ex vivo tissue to in vivo MRI that builds on previous efforts (Goubran et al., 2015; Wisse et al., 2016a). To our knowledge, this is the first such protocol for aligning ex vivo tissue to in vivo MRI in the same subjects to construct a probabilistic atlas based on these subfield boundaries. While past studies have employed group atlases from in vivo MRI-based tracing protocols, or have used a combination of histology and MRI acquired ex vivo (Adler et al., 2016), our method is the first to employ a histologically-driven approach in the same monkeys as those that formed the basis for an in vivo MRI group template. Finally, we use this atlas to address the question of whether subfield volume declines with aging. No past studies have directly compared monkey hippocampal subfield volumes in the same study and we believe that our initial findings further reinforce the idea that hippocampal volumes do not change radically with age in the rhesus macaque.
Using our protocol, which is based on ex vivo Nissl-stained hippocampal post-mortem sections, we show that labels assigned by different raters to the same tissue were largely consistent, suggesting that our method was reliable. While previous work has shown consistency between raters when labeling in vivo MRI images (Dice similarity ranging from 0.75-0.95), MRI-based protocols often take a largely geometric approach to subfield labeling (e.g., drawing supporting lines and boxes on top of images to aid in demarcation (Berron et al., 2017; Entis et al., 2012; Mueller et al., 2007; Pluta et al., 2012). While such approaches are no doubt important, as they support consistent labeling between raters in lieu of clear anatomical boundaries, here we use cytoarchitectural features (e.g., cell body size, orientation, density, and presence of certain interneuron types) to label subfields, with minimal reliance on rules, and demonstrate comparable inter-rater reliability scores (.74-.92). The methods in this study, utilizing actual cellular features of the tissue, are in fact, just as reliable as geometric approaches but importantly, are histologically-driven. In this way, our protocol provides a “ground truth” for identifying hippocampal subfields based on cell density, dendritic orientation, and other features that are only clearly evident using ex vivo Nissl-stained sections.
One caveat with our approach was the relatively low similarity in parasubiculum observed on some sections, and a general trend to lose some reliability as the alveus disappears in the white matter between the entorhinal cortex and subicular regions. For subiculum and presubiculum, our strategy to overcome the lack of clarity in the inferior boundary was to attempt to maintain the distance between the cell body layer and the alveus that was seen before the alveus had disappeared. For parasubiculum, some sections are distinguished by a separate cluster of cell bodies, while some sections are distinguished simply by a lack of organization that occurs between the entorhinal cortex and presubiculum. Sections where parasubiculum is defined by a lack of cellular organization are particularly tricky, and may require additional staining on neighboring sections in order to achieve the accuracy observed in other regions. Overall, however we feel that tissue-based subfield labeling has much to offer hippocampal subfield research, particularly when combined with the registration techniques developed in this manuscript.
Another contribution we make with the data presented here is to refine a set of methods that address the computational complexity required to make tissue-based subfield labeling useful to in vivo MRI-based research. Here, we propose a set of methods for aligning tissue to in vivo MRI and combining the results into a probabilistic atlas. Our methods draw heavily from previous work, which had largely addressed the problem of registering histology to MRI (Adler et al., 2014), or creating group-templates from high-resolution images (Iglesias et al., 2015). Our primary advance in this domain was to modify these approaches, and in some cases, simplify them, in order to make a pipeline robust enough that parameters did not need to be changed for different subjects, and efficient enough to run on a high-end PC rather than a cluster or server. Using these methods, we observed overall low error, roughly 1-2 voxels. One manual aspect of our otherwise automated pipeline is the affine registration step between the histology and the MRI. Although multiple implementations of automated registration were considered for this step, the results were unsatisfactory, and thus, this manual step remained in our pipeline. We believe the relatively low resolution and contrast of our structural MRIs may be responsible for the ineffectiveness of the automated methods, and thus, in the future we plan to address this issue with higher-contrast imaging sequences (e.g., Ekstrom et al., 2009). Nonetheless, the overall low error observed here suggests that we have achieved accurate registration despite the use of manual steps and low resolution of the 1.5T MRI.
Having developed a histologically-based protocol for identifying primate subfields on MRIs, we then use our methods to address whether changes in subfield volume accompany normal aging. This is an important issue to address as subfield-specific functional changes have been shown to occur with aging (Thome et al., 2016). We note that comparing young and old brains based on combined MRI and histology, particularly healthy ones in the same subjects, is unlikely to be ethically possible in humans. Thus, such work with non-human primates fills an important void that would otherwise be unlikely to be filled with human work. Interestingly previous work in the same monkeys had suggested that total hippocampal volume did not decrease with age (Shamy et al., 2006). Studies in humans utilizing in vivo subfield labeling, however, have indicated inverse correlations between age and both whole hippocampal, and subfield volume using MRI (Mueller et al., 2007; Voineskos et al., 2015), however, effect sizes were small. Thus, while we show tentative evidence that hippocampal subfield volume does not change in the aging macaque, further work will need to incorporate significantly more subjects in order to address the subtle changes in subfield volume that may be expected based on studies done in humans.
With future work, we hope to both extend the functionality of the current methods and address some of the technical complications of the current technique. First, we hope to continue adding subjects to the atlas in order to more accurately reflect the degree of variability between subjects, and increase statistical power. Second, we hope to integrate higher resolution MRI images, including T2 weighted coronal images, which most often serve as the basis for in vivo tracing of human subjects. If reliable features in T2 images can be shown to coincide with cytoarchitectural landmarks, this would constitute a significant advance in subfield demarcation and provide a clear path of translation between our macaque dataset and the human hippocampus. Recent efforts linking cytoarchitectural features to MRI intensity gradients in human patient populations have been fruitful. For example, (Wisse et al., 2016a) has demonstrated a correspondence between hypointense strips in the hippocampal head, visible in T2 images, with anatomical features of region CA3. Thus, efforts in the nonhuman primate domain, across the lifespan of healthy animals, should complement and help to validate research efforts in humans. Another area for future development is to better quantify individual variability in subfield boundary locations. Future work will attempt to disentangle error in subfield demarcation generated from the histology to MRI registration step, from those generated from the template creation process step, enabling us to construct better estimates for the degree of individual variability in subfield boundary locations. Finally, we hope to move toward automated segmentation techniques, and to test the ability of these methods to capture individual variability.
Another potential area to which our method could be applied is to human subfield demarcation. There are strong similarities between the monkey and human hippocampus and most aspects of hippocampal neuroanatomy, including relative cell density, cytoarchitecture, chemoarchitecture, and neural connectivity, appear to be conserved (Amaral and Lavenex, 2007; Amaral et al., 2007; Ding, 2013; Ding et al., 2010; Lim et al., 1997). Therefore, developing a framework that may improve the accuracy of human subfield labeling based on data drawn from nonhuman primates would be an important advance. Developing such a framework will require careful design decisions. For instance, multi-atlas group-voting based techniques are promising (Pipitone et al., 2014; Yushkevich et al., 2010), however the methods do not lend themselves to translation between species. One approach that we feel offers hope is the Bayesian framework presented by Van Leemput et al. (2009). This method relies on a spatial-mesh prior which specifies the probability of subfield identity as a function of location. The prior is updated with a likelihood function that is based on the image intensity values found in each region. The clear dichotomy of spatial location verses intensity values offers a solution to transferring information between species. While the exact spatial relationships between subfields in rhesus macaque hippocampus is not identical to those in humans, the cellular and chemical makeup of these regions are highly similar (Amaral and Lavenex, 2007; Ding, 2013), and thus, MRI intensity values within each subregion will also likely show considerable similarity between species. Thus, by incorporating the histologically-based subfield intensity values from the current methodology, the Bayesian framework offers a path to translation between nonhuman primates and humans for automated subfield classification.
In summary, our approach provides a novel manual demarcation protocol for Nissl-stained sections and the methods necessary to apply manual tissue-based labels to in vivo MRI in a mostly automated fashion. Using these methods, we conclude that subfield volumes are unlikely to show large changes across the lifespan of the rhesus macaque. Additionally, our method produces strong correspondence between true subfield identity and MRI intensity values, information that will be crucial in refining both manual and automated methods in humans. This approach enables a means to standardize subfield labeling protocols and refine automated labeling methods, potentially resulting in more consistency across laboratories.
Supplementary Material
Supplemental Figure 2. Examples of individual registration quality. A. Overlay of histology and MRI after 3D rigid registration. B. Overlay of histology and MRI after 2D and 3D non-rigid registration.
Supplemental Figure 3. Subfield volume comparison between aged and young rhesus macaques. Group-space subfield volumes for young and aged animals. Mann-Whitney U-tests were conducted between age groups for each subfield. Results indicated no significant differences in subfield volumes between age groups (p>.39, corrected).
Acknowledgments
We’d like to thank Mike Buonocore for his assistance with MRI imaging of these animals, and David Amaral for providing calretinin-stained tissue sections for our anatomical protocol development. Supported by: McKnight Brain Research Foundation, NIH grants R01 AG003376, P51 RR000169, R01 NS076856.
References
- Adler DH, Ittyerah R, Pluta J, Pickup S, Liu W, Wolk DA, Yushkevich PA. Probabilistic Atlas of the Human Hippocampus Combining Ex Vivo MRI and Histology. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016: 19th International Conference; Athens, Greece. October 17-21, 2016; Springer International Publishing; 2016. pp. 63–71. Proceedings, Part III. Cham. [Google Scholar]
- Adler DH, Pluta J, Kadivar S, Craige C, Gee JC, Avants BB, Yushkevich PA. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage. 2014;84:505–23. doi: 10.1016/j.neuroimage.2013.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral D, Lavenex P. Hippocampal Neuroanatomy. In: Andersen P, Morris R, Amaral D, Bliss T, O’Keefe J, editors. The Hippocampus Book. Oxford University Press Inc.; 2007. [Google Scholar]
- Amaral DG, Scharfman HE, Lavenex P. The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies) Prog Brain Res. 2007;163:3–22. doi: 10.1016/S0079-6123(07)63001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–44. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A, Kirwan CB, Miller M, Stark CE. Pattern separation in the human hippocampal CA3 and dentate gyrus. Science. 2008;319(5870):1640–2. doi: 10.1126/science.1152882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berron D, Vieweg P, Hochkeppler A, Pluta JB, Ding SL, Maass A, Luther A, Xie L, Das SR, Wolk DA, et al. A protocol for manual segmentation of medial temporal lobe subregions in 7 Tesla MRI. Neuroimage Clin. 2017;15:466–482. doi: 10.1016/j.nicl.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckmaster PS, Amaral DG. Intracellular recording and labeling of mossy cells and proximal CA3 pyramidal cells in macaque monkeys. J Comp Neurol. 2001;430(2):264–81. doi: 10.1002/1096-9861(20010205)430:2<264::aid-cne1030>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- de Toledo-Morrell L, Morrell F. Electrophysiological markers of aging and memory loss in rats. Ann N Y Acad Sci. 1985;444:296–311. doi: 10.1111/j.1749-6632.1985.tb37598.x. [DOI] [PubMed] [Google Scholar]
- Ding SL. Comparative anatomy of the prosubiculum, subiculum, presubiculum, postsubiculum, and parasubiculum in human, monkey, and rodent. J Comp Neurol. 2013;521(18):4145–62. doi: 10.1002/cne.23416. [DOI] [PubMed] [Google Scholar]
- Ding SL, Haber SN, Van Hoesen GW. Stratum radiatum of CA2 is an additional target of the perforant path in humans and monkeys. Neuroreport. 2010;21(4):245–9. doi: 10.1097/WNR.0b013e328333d690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SL, Van Hoesen GW. Borders, extent, and topography of human perirhinal cortex as revealed using multiple modern neuroanatomical and pathological markers. Hum Brain Mapp. 2010;31(9):1359–79. doi: 10.1002/hbm.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SL, Van Hoesen GW. Organization and Detailed Parcellation of Human Hippocampal Head and Body Regions Based on a Combined Analysis of Cyto-and Chemoarchitecture. J Comp Neurol. 2015;523(15):2233–53. doi: 10.1002/cne.23786. [DOI] [PubMed] [Google Scholar]
- Ekstrom AD, Bazih AJ, Suthana NA, Al-Hakim R, Ogura K, Zeineh M, Burggren AC, Bookheimer SY. Advances in high-resolution imaging and computational unfolding of the human hippocampus. Neuroimage. 2009;19:19. doi: 10.1016/j.neuroimage.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entis JJ, Doerga P, Barrett LF, Dickerson BC. A reliable protocol for the manual segmentation of the human amygdala and its subregions using ultra-high resolution MRI. Neuroimage. 2012;60(2):1226–35. doi: 10.1016/j.neuroimage.2011.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30(9):1323–41. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MS, Kraemer GW, Shelton SE, Baker E, Kalin NH, Uno H. Gender differences in brain volume and size of corpus callosum and amygdala of rhesus monkey measured from MRI images. Brain Res. 2000;852(2):263–7. doi: 10.1016/s0006-8993(99)02093-4. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Jack CR. HarP: the EADC-ADNI Harmonized Protocol for manual hippocampal segmentation. A standard of reference from a global working group. Alzheimers Dement. 2015;11(2):107–10. doi: 10.1016/j.jalz.2014.05.1761. [DOI] [PubMed] [Google Scholar]
- Goubran M, de Ribaupierre S, Hammond RR, Currie C, Burneo JG, Parrent AG, Peters TM, Khan AR. Registration of in-vivo to ex-vivo MRI of surgically resected specimens: a pipeline for histology to in-vivo registration. J Neurosci Methods. 2015;241:53–65. doi: 10.1016/j.jneumeth.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Knierim JJ, Moser EI. Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron. 2004;44(4):581–4. doi: 10.1016/j.neuron.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage. 2015;115:117–37. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchner GA, Deutsch GK, Zeineh M, Dougherty RF, Saranathan M, Rutt BK. Hippocampal CA1 apical neuropil atrophy and memory performance in Alzheimer’s disease. Neuroimage. 2012;63(1):194–202. doi: 10.1016/j.neuroimage.2012.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Blume HW, Madsen JR, Saper CB. Connections of the hippocampal formation in humans: I. The mossy fiber pathway. J Comp Neurol. 1997;385(3):325–51. doi: 10.1002/(sici)1096-9861(19970901)385:3<325::aid-cne1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Stables L, Du AT, Schuff N, Truran D, Cashdollar N, Weiner MW. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4T. Neurobiol Aging. 2007;28(5):719–26. doi: 10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunuebel JP, Knierim JJ. CA3 retrieves coherent representations from degraded input: direct evidence for CA3 pattern completion and dentate gyrus pattern separation. Neuron. 2014;81(2):416–27. doi: 10.1016/j.neuron.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos GHX-F, Petrides M, Toga AW. The Rhesus Monkey Brain. Academic Press; 2009. [Google Scholar]
- Pipitone J, Park MTM, Winterburn J, Lett TA, Lerch JP, Pruessner JC, Lepage M, Voineskos AN, Chakravarty MM, Initiative AsDN Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014;101:494–512. doi: 10.1016/j.neuroimage.2014.04.054. [DOI] [PubMed] [Google Scholar]
- Pluta J, Yushkevich P, Das S, Wolk D. In vivo analysis of hippocampal subfield atrophy in mild cognitive impairment via semi-automatic segmentation of T2-weighted MRI. J Alzheimers Dis. 2012;31(1):85–99. doi: 10.3233/JAD-2012-111931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski E, Stebbins GT, Barnes CA, Murphy CM, Stoub TR, George S, Ferrari C, Shah RC, deToledo-Morrell L. Age-related changes in parahippocampal white matter integrity: a diffusion tensor imaging study. Neuropsychologia. 2012;50(8):1759–65. doi: 10.1016/j.neuropsychologia.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamy JL, Buonocore MH, Makaron LM, Amaral DG, Barnes CA, Rapp PR. Hippocampal volume is preserved and fails to predict recognition memory impairment in aged rhesus monkeys (Macaca mulatta) Neurobiol Aging. 2006;27(10):1405–15. doi: 10.1016/j.neurobiolaging.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Stokes J, Kyle C, Ekstrom AD. Complementary Roles of Human Hippocampal Subfields in Differentiation and Integration of Spatial Context. J Cogn Neurosci. 2015;27(3):549–559. doi: 10.1162/jocn_a_00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoub TR, Barnes CA, Shah RC, Stebbins GT, Ferrari C, deToledo-Morrell L. Age-related changes in the mesial temporal lobe: the parahippocampal white matter region. Neurobiol Aging. 2012;33(7):1168–76. doi: 10.1016/j.neurobiolaging.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoub TR, Detoledo-Morrell L, Dickerson BC. Parahippocampal white matter volume predicts Alzheimer’s disease risk in cognitively normal old adults. Neurobiol Aging. 2014;35(8):1855–61. doi: 10.1016/j.neurobiolaging.2014.01.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthana NA, Donix M, Wozny DR, Bazih A, Jones M, Heidemann RM, Trampel R, Ekstrom AD, Scharf M, Knowlton B, et al. High-resolution 7T fMRI of Human Hippocampal Subfields during Associative Learning. Journal of Cognitive Neuroscience. 2015;27(6):1194–1206. doi: 10.1162/jocn_a_00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome A, Gray DT, Erickson CA, Lipa P, Barnes CA. Memory impairment in aged primates is associated with region-specific network dysfunction. Mol Psychiatry. 2016;21(9):1257–62. doi: 10.1038/mp.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges J, Gordon TP, Mcclure HM, Hall EC, Peters A. Survival Rate and Life-Span of Rhesus-Monkeys at the Yerkes-Regional-Primate-Research-Center. American Journal of Primatology. 1988;15(3):263–273. doi: 10.1002/ajp.1350150308. [DOI] [PubMed] [Google Scholar]
- van Dijk RM, Huang SH, Slomianka L, Amrein I. Taxonomic Separation of Hippocampal Networks: Principal Cell Populations and Adult Neurogenesis. Front Neuroanat. 2016;10:22. doi: 10.3389/fnana.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19(6):549–57. doi: 10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage. 2009;45(1 Suppl):S61–72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Vieweg P, Stangl M, Howard LR, Wolbers T. Changes in pattern completion–a key mechanism to explain age-related recognition. Cortex. 2015;64:343–51. doi: 10.1016/j.cortex.2014.12.007. LID - 10.1016/j.cortex.2014.12.007 [doi] LID - S0010-9452(14)00410-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos AN, Winterburn JL, Felsky D, Pipitone J, Rajji TK, Mulsant BH, Chakravarty MM. Hippocampal (subfield) volume and shape in relation to cognitive performance across the adult lifespan. Hum Brain Mapp. 2015;36(8):3020–37. doi: 10.1002/hbm.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Ikonen S, Gallagher M, Eichenbaum H, Tanila H. Age-associated alterations of hippocampal place cells are subregion specific. J Neurosci. 2005;25(29):6877–86. doi: 10.1523/JNEUROSCI.1744-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse LE, Adler DH, Ittyerah R, Pluta JB, Robinson JL, Schuck T, Trojanowski JQ, Grossman M, Detre JA, Elliott MA, et al. Comparison of In Vivo and Ex Vivo MRI of the Human Hippocampal Formation in the Same Subjects. Cereb Cortex. 2016a doi: 10.1093/cercor/bhw299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse LE, Daugherty AM, Olsen RK, Berron D, Carr VA, Stark CE, Amaral RS, Amunts K, Augustinack JC, Bender AR, et al. A harmonized segmentation protocol for hippocampal and parahippocampal. 2016b doi: 10.1002/hipo.22671. LID - 10.1002/hipo.22671 [doi]. Hippocampus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Lacy JW, Stark SM, Albert MS, Gallagher M, Stark CE. Pattern separation deficits associated with increased hippocampal CA3 and dentate gyrus activity in nondemented older adults. Hippocampus. 2011;21(9):968–79. doi: 10.1002/hipo.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Amaral RS, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, Bocchetta M, Burggren AC, Carr VA, Chakravarty MM, et al. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. Neuroimage. 2015a;111:526–41. doi: 10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Pluta JB, Wang H, Xie L, Ding SL, Gertje EC, Mancuso L, Kliot D, Das SR, Wolk DA. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp. 2015b;36(1):258–87. doi: 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage. 2010;53(4):1208–1224. doi: 10.1016/j.neuroimage.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Bookheimer SY. Application of cortical unfolding techniques to functional MRI of the human hippocampal region. Neuroimage. 2000;11(6)(Pt 1):668–83. doi: 10.1006/nimg.2000.0561. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Unfolding the human hippocampus with high resolution structural and functional MRI. Anat Rec. 2001;265(2):111–20. doi: 10.1002/ar.1061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 2. Examples of individual registration quality. A. Overlay of histology and MRI after 3D rigid registration. B. Overlay of histology and MRI after 2D and 3D non-rigid registration.
Supplemental Figure 3. Subfield volume comparison between aged and young rhesus macaques. Group-space subfield volumes for young and aged animals. Mann-Whitney U-tests were conducted between age groups for each subfield. Results indicated no significant differences in subfield volumes between age groups (p>.39, corrected).


