Fig. 1.
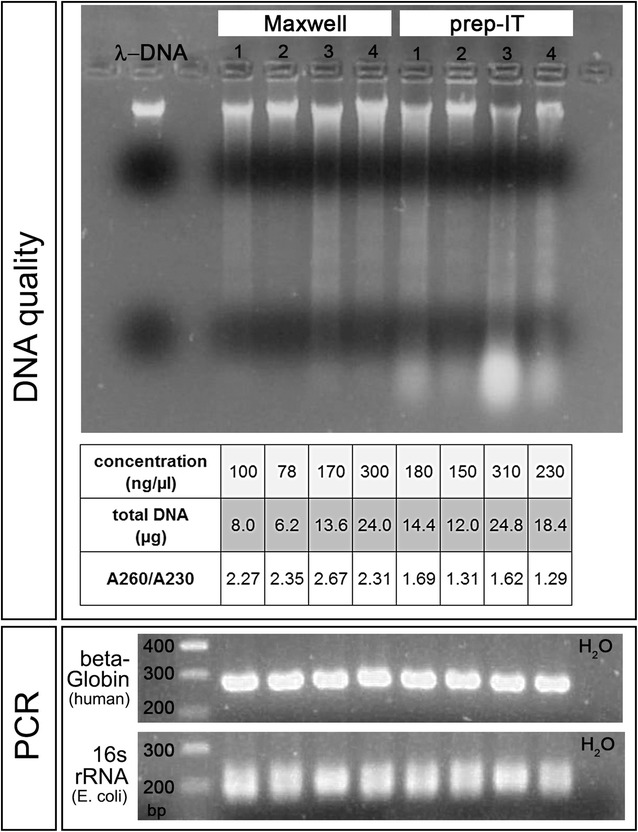
Comparison of quality and bacterial contamination of DNA extracted using two different methods. DNA from four saliva samples (number 1–4) was isolated automatically (Maxwell) or manually (prep-IT). DNA quality was assessed by agarose gel electrophoresis. High-quality DNA is represented by a lambda DNA (300 ng) probe (first lane). DNA quantity was measured by fluorometric assessment (Quantus) and the purity of the DNA was quantified by photometric analysis of absorbance at 260 and 230 nm (A260/A230). PCR amplification of human beta-Globin DNA and bacterial 16s rRNA was used to determine differences in bacterial DNA contamination. In the first lane, a 100 base pairs (bp) DNA marker determines the size of the amplified fragments
