Figure 1.
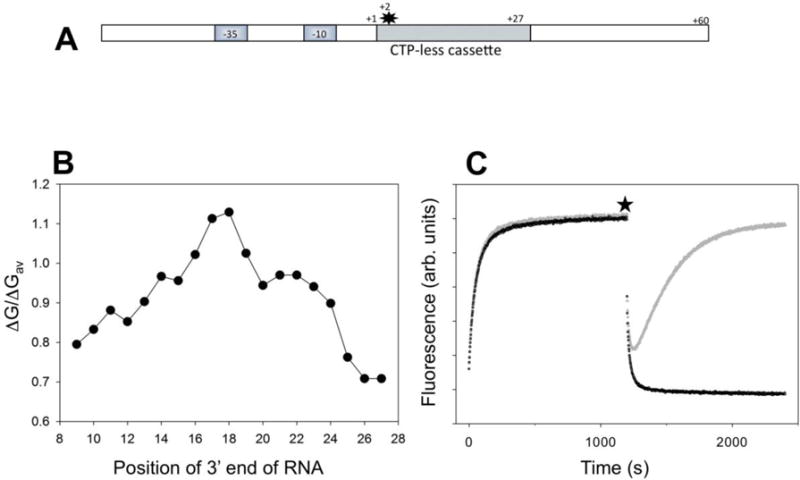
(A) Design of fluorescent construct (P27 promoter) for real-time detection of RNAP backtracking. The star denotes Cy3 label at the +2 position of the nontemplate strand. (B) Calculated relative duplex stabilities (ratio of duplex free energy of the 9 bp hybrid to the average of free energies of all possible sequence combinations for 9 bp long duplexes) of the 9 bp RNA:DNA hybrids along the sequence of the first 27 transcribed nucleotides of P27. (C) Fluorescence intensity changes of P27. At time 0 the P27 was added to a solution of RNAP in a fluorometer cuvette. At the time point marked by a star, the transcription was started by adding ATP, UTP, GTP and heparin (grey) or ATP, UTP, GTP, CTP and heparin (black).
