Fig. 2.
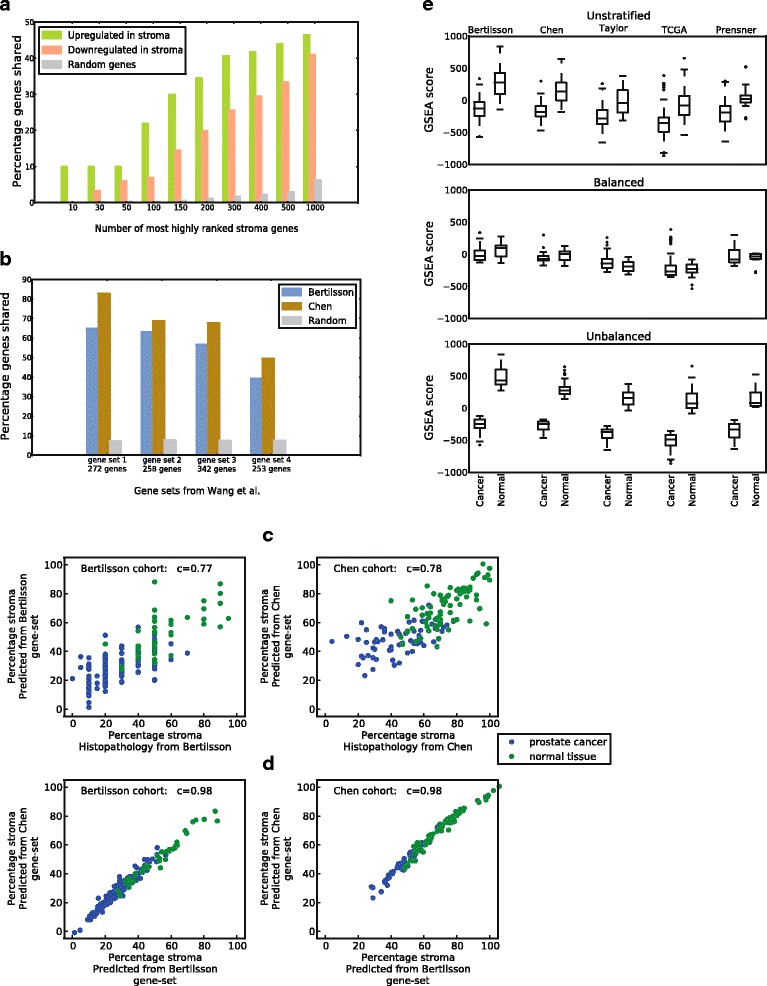
Robust assessment of stroma content in cohorts where no histopathology is available. a Overlap between up- and downregulated stroma genes in the gene-sets from Bertilsson and Chen for various numbers of the N top-ranked stroma genes. The random numbers of shared genes are the average over 50 random gene selections for each N. b Overlap of prostate stroma gene sets from Wang et al. [21] with stroma gene-sets from Bertilsson and Chen. c Pearson correlation (c) between predicted stroma percentage from GSEA and histopathological determined stroma percentage in the cohorts from Bertilsson and Chen. d Pearson correlation (c) between stroma percentage predicted by gene-sets from Bertilsson and Chen in each of their respective cohorts (bottom). e Bias towards higher GSEA stroma scores in normal compared to cancer samples present in all unstratified cohorts from the five-study-cohort. Dividing samples into balanced and unbalanced datasets minimizes and maximizes, respectively, the stroma bias between cancer and normal samples. A difference in the average overall GSEA score between the cohorts is also evident in the figure
