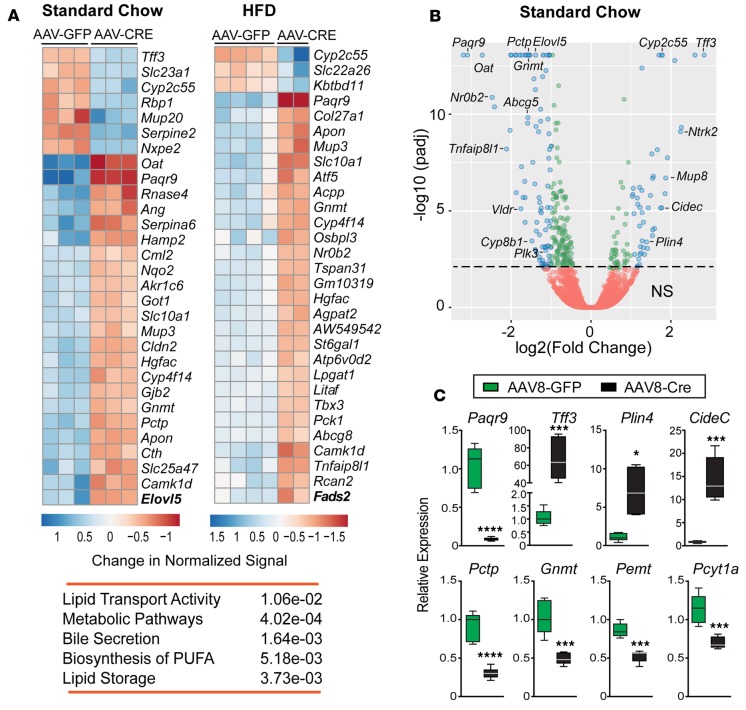
Figure 4. Lipid storage and phospholipid metabolic pathways are affected in Lrh-1AAV8-Cre mice.
(A) Top 30 differentially expressed genes (DEGs) and significant KEGG pathways in livers of Lrh-1AAV8-Cre male mice fed standard diet (SD) or high-fat diet (HFD). (B) Volcano plot of DEGs between Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice fed SD, with DEGs of adjusted P < 0.01 (green circles) and those with adjusted P < 0.01 and absolute log2 fold change >1.0 (blue circles). Annotated genes include established LRH-1 targets (Cyp8b1, Nr0b2, and Plk3), top DEGs (Paqr9 and Tff3), and DEGs associated with lipid droplet size and formation (Plin4 and Cidec) and phospholipid biosynthesis (Pctp and Gnmt). (C) Validation of select DEGs by qPCR analysis in livers from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice fed SD (Lrh-1AAV8-GFP, n = 5, and Lrh-1AAV8-Cre, n = 6). Error bars represent ± SEM. For box-and-whisker plots, maximum and minimum values are shown with median. *P < 0.05, ***P < 0.001, ****P < 0.0001, unpaired Student’s t test.