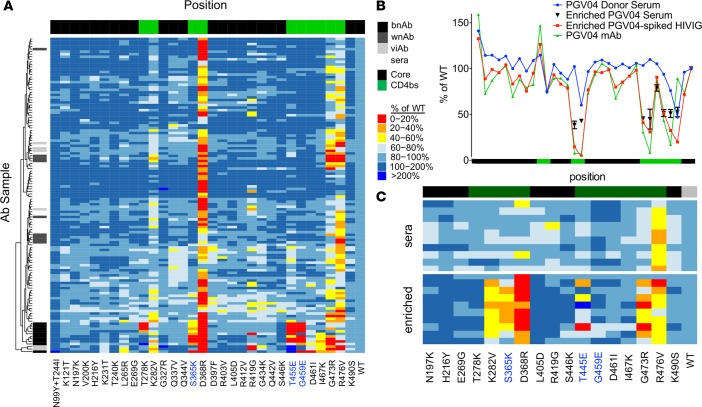
Figure 3. Polyclonal sera epitope-mapping results.
(A) Heatmap comparing epitope-mapping data from polyclonal sera from Boston area (Ragon Institute) HIV-infected subjects positive for YU2 gp120 core-specific antibodies clustered together with the mAb epitope maps. The vertical color bar indicates sample type: pAb samples are shown in white (n = 99), bnAbs in black (n = 10), vaccine-induced weakly neutralizing antibodies (viAbs, n = 6) in light gray, and infection-induced weakly neutralizing antibodies (wnAbs, n = 9) in dark gray. (B) Serum from a donor with a known bnAb (PGV04) was evaluated for binding to the epitope-mapping panel. The ability of the STG mutant to deplete core-specific antibodies without broad neutralization and to facilitate identification of the presence of CD4bs antibodies with broad neutralization was determined for the PGV04 donor serum and for PGV04-spiked HIVIG. Error bars, shown only for the enriched PGV04 serum, indicate SD observed between duplicate measurements. (C) Heatmap of polyclonal sera samples from a Tanzanian cohort (n = 10) before and after enrichment of potential bnAbs using the STG mutant-based depletion. Horizontal color bars indicate the class of core: core variants with substitutions made in CD4bs residues are indicated in green, core variants with substitutions made in other sites on the core are indicated in black.