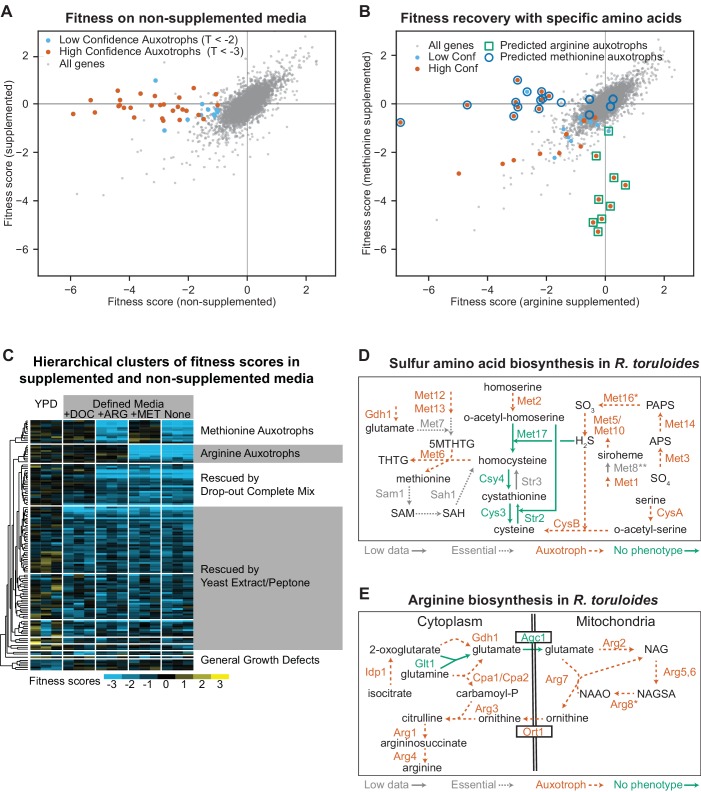
Figure 2. Confirmation of amino acid biosynthetic genes with high-throughput fitness experiments.
(A) Fitness scores for 6,558 genes in media with and without amino acid supplementation (drop-out complete mix). Gene fitness scores are log ratios of final versus starting abundance averaged over multiple barcoded insertions per gene across three biological replicates. Genes that had consistently different enrichment scores between treatments (∆F > 1, |T| statistic >3) are highlighted and represent genes for which mutant strains are auxotrophic for one or more amino acids, nucleotides, or vitamins present in the drop-out complete mixture. (B) Fitness scores in media supplemented with arginine or methionine. Highlighted genes are the same as highlighted in (A). Deletion strains for circled or boxed genes are auxotrophic for methionine or arginine, respectively, in S. cerevisiae or A. nidulans. See Supplementary file 2 for full fitness data. (C) Hierarchical clusters of fitness scores in supplemented and non-supplemented media. Fitness scores for each biological replicate versus its Time 0 replicate for genes with a consistent fitness defect (F < −1, T < −3) in one or more of the following conditions: Yeast extract/Peptone/Dextrose media (YPD) or defined media (DM, composed of yeast nitrogen base plus glucose) with or without the following supplements: (+DOC), arginine (+ARG), or methionine (+MET). (D) Sulfur amino acid biosynthesis in R. toruloides as inferred from fitness experiments. CysA/CysB are named according to their A. nidulans orthologs, all others by orthologs in S. cerevisiae. Auxotrophic mutants had F < −1 in non-supplemented media (DM) and T < −3 in DM versus supplemented media (DOC). Multiple insertions were mapped in STR3, suggesting non-essentiality, but strain abundance was too low to estimate fitness in BarSeq data. *MET16 had fitness scores that clustered with the other auxotrophic mutants, but TDM-DOC was −2.7. **Fitness scores for insertions in MET8 were not inconsistent with auxotrophy, but only two insertions were abundant enough to be tracked. 5MTHTG: 5-methyltetrahydropteroyltri-L-glutamate, THTG: tetrahydropteroyltri-L-glutamate, SAM: S-adenosyl-L-methionine, SAH: S-adenosyl-homocysteine, APS: adenylyl-sulfate, PAPS: 3'-phosphoadenylyl-sulfate. (E) Arginine biosynthesis in R. toruloides as inferred from fitness experiments. *ARG8 had fitness scores that clustered with the other auxotrophic mutants, but TDM-DOC was −2.9. NAG: N-acetylglutamate, NAGSA: N-acetylglutamate semialdehyde, NAAO: N-alpha-acetylornithine. The following figure supplements are available for Figure 2.