Figure 3. Genes with fitness defects on fatty acids.
(A) Heatmap of fitness scores for R. toruloides genes with predicted roles in beta-oxidation of fatty acids. Enzyme classes and predicted locations were inferred from homologous proteins in Ustilago maydis as reported by Camões et al. (Camões et al., 2015). See Supplementary file 2 for full fitness data. (B) Log2 optical density ratio for single deletion mutants versus the YKU70∆ control strain at mid-log phase on 1% oleic acid as carbon source are plotted against the fitness scores for each gene from BarSeq experiments on 1% oleic acid. (C) Log2 optical density ratio for single deletion mutants versus the YKU70∆ control strain at mid-log phase on 1% ricinoleic acid as carbon source are plotted against the fitness scores for mutants in each gene from BarSeq experiments on 1% ricinoleic acid. See Supplementary file 2 for a statistical summary for all strains shown in (B) and (C), including P values and effect sizes. The following figure supplements are available for Figure 3.
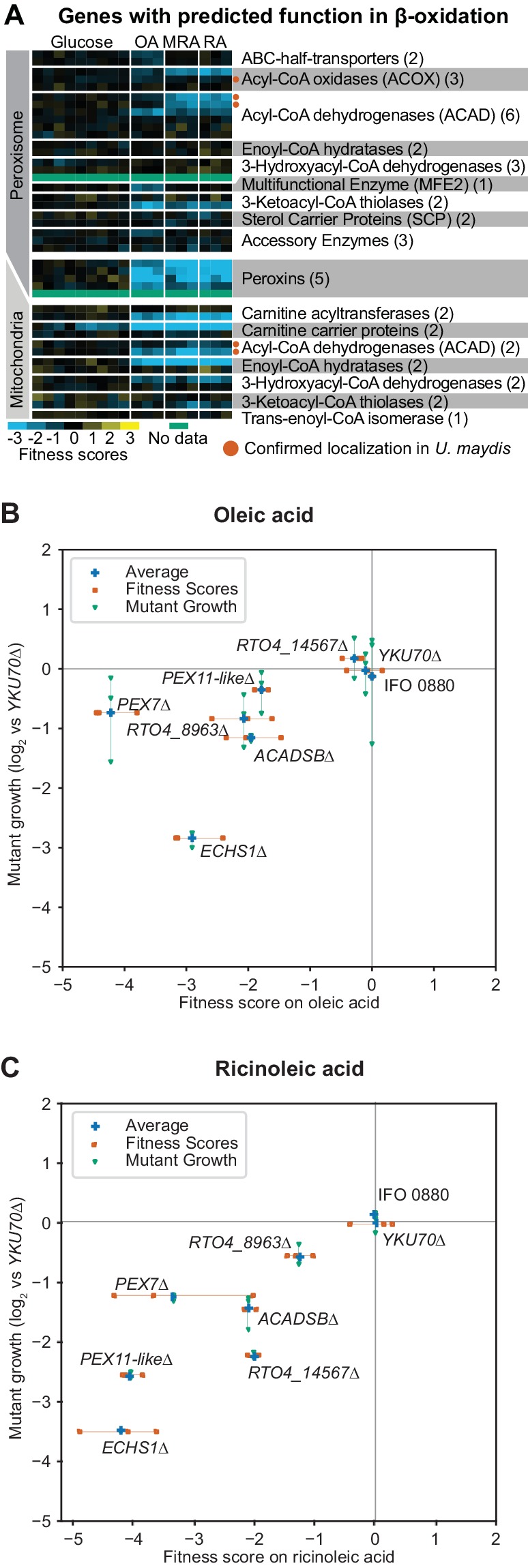
Figure 3—figure supplement 1. K-means clusters of fitness scores for 129 genes for which mutants have specific fitness defects on fatty acids.
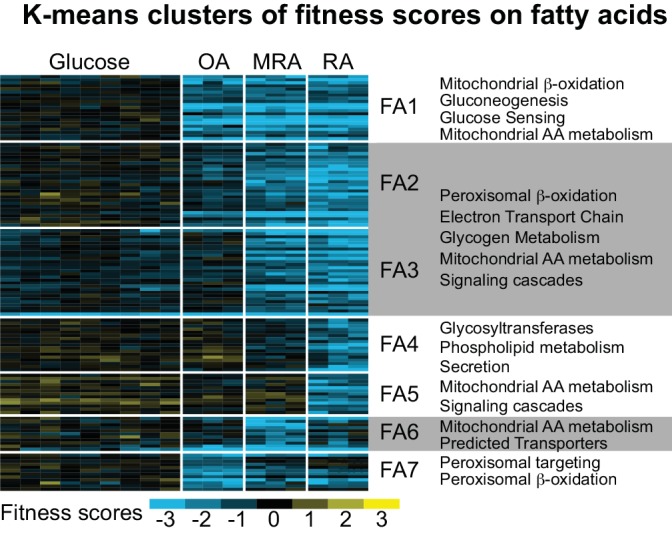
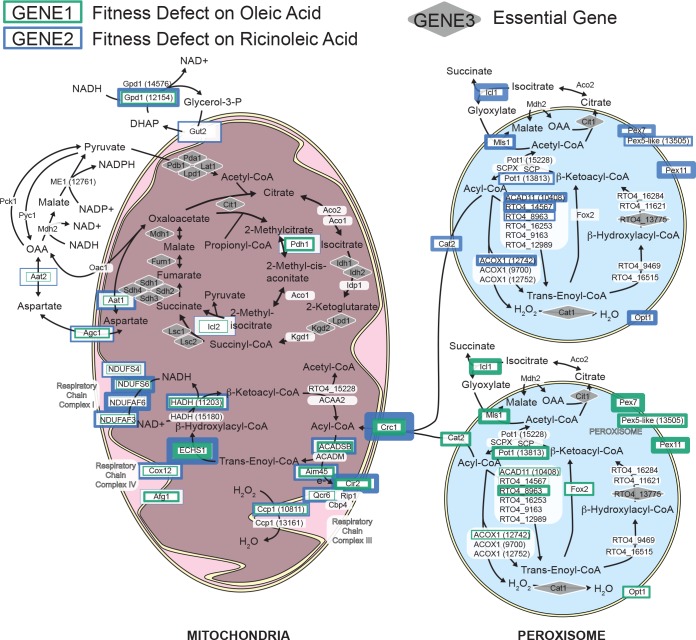
Figure 3—figure supplement 2. Model for beta-oxidation of fatty acids in R. toruloides.
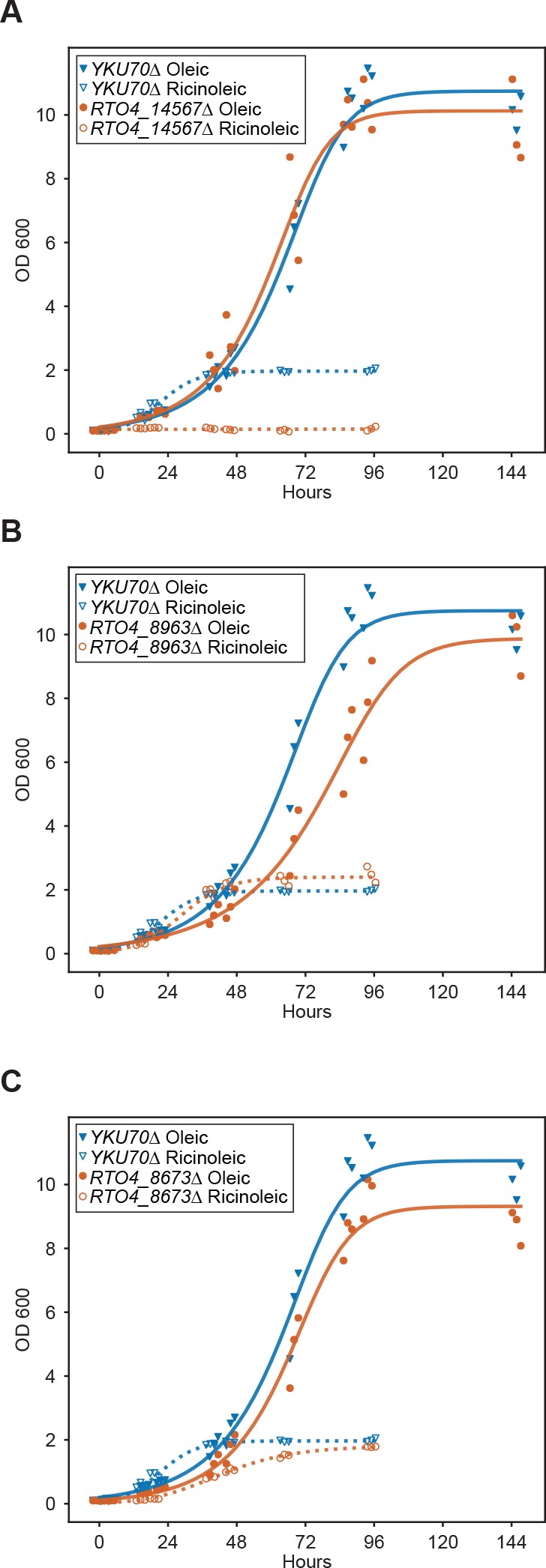
Figure 3—figure supplement 3. Extended growth curves for deletion mutants on fatty acids.