Abstract
Background
Reported per-patient costs of Clostridium difficile infection (CDI) vary by two orders of magnitude among different hospitals, implying that infection control officers need precise, local analyses to guide rational decision-making between interventions.
Objective
We sought to comprehensively estimate changes in length of stay (LOS) attributable to CDI at one urban tertiary-care facility using only data automatically extractable from the electronic medical record (EMR).
Methods
We performed a retrospective cohort study of 171,938 visits spanning a 7-year period. 23,968 variables were extracted from EMR data recorded within 24 hours of admission to train elastic net regularized logistic regression models for propensity score matching. To address time-dependent bias (reverse causation), we separately stratified comparisons by time-of-infection and fit multistate models.
Results
The estimated difference in median LOS for propensity-matched cohorts varied from 3.1 days (95% CI, 2.2–3.9) to 10.1 days (95% CI, 7.3–12.2) depending on the case definition; however, dependency of the estimate on time-to-infection was observed. Stratification by time to first positive toxin assay, excluding probable community-acquired infections, showed a minimum excess LOS of 3.1 days (95% CI, 1.7–4.4). Under the same case definition, the multistate model averaged an excess LOS of 3.3 days (95% CI, 2.6–4.0).
Conclusions
Two independent time-to-infection adjusted methods converged on similar excess LOS estimates. Changes in LOS can be extrapolated to a marginal dollar costs by multiplying by average costs of an inpatient-day. Infection control officers can leverage automatically extractable EMR data to estimate costs of CDI at their own institution.
Keywords: Clostridium difficile, attributable cost, clinical informatics, length of stay, machine learning
Introduction
Clostridium difficile infection (CDI) is the most frequently reported healthcare-associated infection (HAI) in the US1 and the major infective cause of nosocomial diarrhea in developed countries,2 incurring billions of dollars in excess medical costs per year.3 Estimates of the per-patient cost of CDI have varied from $2,871 to $122,318 due to differences in methodology, patient inclusion criteria, and regional costs.4–6 Given the high hospital-to-hospital variability of these costs,7,8 infection control officers, hospital administrators, and clinicians would benefit from estimates tailored to their particular population and healthcare practices. Concretely defining the potential economic savings of CDI prevention would empower stakeholders to prudently choose among the many available validated interventions.9,10
Measuring costs within healthcare systems is notoriously difficult, as many hospitals do not have access to itemized reimbursement data linked to medical records.11 Even institutions that have informatics retrospectively linking these data have relied on the curation of select variables and chart review to estimate attributable CDI cost.12–14 Nevertheless, electronic medical record (EMR) systems are used by most first-world acute care facilities.15,16 Part of the rationale for these systems is that hospitals may leverage EMR data for optimal decision-making by inferring causal relationships from raw observations during routine care.17–19 An analysis based on automatically extractable data from an EMR that quantifies preventable hospital costs, such as those attributable to an HAI like CDI, would be of great value in building a continuously learning healthcare system.20 EMRs contain many structured fields relevant to this analysis, including: diagnosis codes and lab results demonstrating onset of HAIs; thousands of variables for procedures, problems, and medications that can serve as covariates for adjustment in observational studies; and importantly, the length of stay (LOS) for each visit, which is the primary contributor to excess costs for most HAIs, including CDI.3,21,22
The goal of this study was to generate a robust estimate of local cost associated with CDI using data that are automatically extractable from a typical EMR. We use all available structured data recorded within 24 hours of admission in the EMR—including over 20,000 variables, such as medications reported and administered, abnormal lab values, and problem list entries—to build fully data-driven models for CDI risk using a machine learning algorithm, avoiding the potential bias of preselected covariates and manual chart review. CDI risk models trained on uncurated data from EMRs have already outperformed models that only incorporate variables for known risk factors, indicating that CDI risk may be nuanced in particular care settings.23 We then use these trained CDI risk models for propensity score matching, which allows estimation of changes in LOS associated with CDI. Most previous studies of CDI cost do not account for the possibility that longer LOS increases the risk of CDI, i.e., reverse causation, and therefore likely overestimate the cost of CDI.7,24 To adjust for this, we stratify our analysis by the time of CDI diagnosis to find the change in LOS conditional on minimal prior exposure to the hospital environment. Finally, we compare these results to a multistate model of competing time-dependent risks between discharge and the onset of CDI.
Methods
Data Source
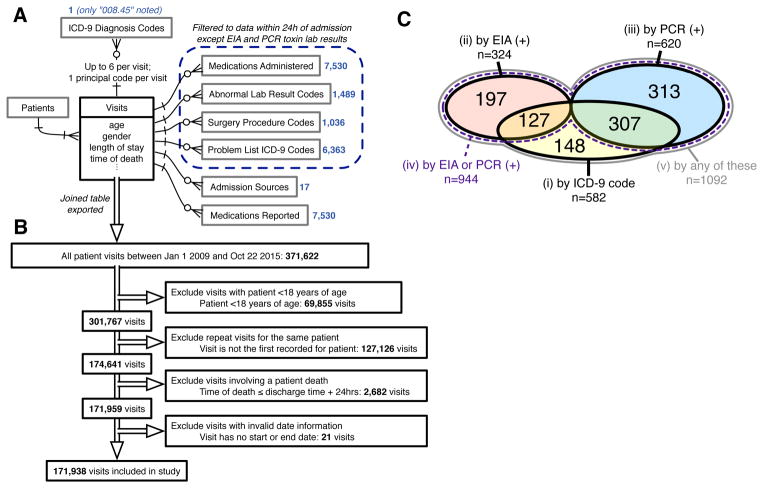
This study was conducted at The Mount Sinai Hospital, a 1,171-bed tertiary care hospital in New York, NY. Records of warehoused adult inpatient EMR visit data were de-identified using the HIPAA Safe Harbor method, 45 CFR §164.514(b)(2). Data was collected on demographics, LOS, time of death, admission sources, reported medications, and the presence of a “008.45” ICD-9 principal or secondary visit diagnosis code denoting “Intestinal infection due to Clostridium difficile.” Furthermore, all records of medications administered, abnormal lab results, surgery procedure codes, or problem list ICD-9 codes within the first 24 hours after admission were collected as Boolean variables (presence or absence). All variables that were uniform across the study population were dropped from the dataset. The relationships between collected data elements are summarized in Figure 1A. This study was approved by Mount Sinai’s Institutional Review Board as exempt research.
Figure 1. Data Sources, Inclusion/Exclusion Criteria, and Cohort Sizes Before Matching.
A, entity-relationship diagram for all EMR data used to generate models of CDI propensity, using Information Engineering notation.44 Boxes represent tables of entities with any directly associated attributes (fields) listed below; single lines represent relationships, with arrowheads indicating the cardinality of each side of the relationship; crow’s foot arrowhead with circle represents “zero or more”; crow’s foot arrowhead with cross-stroke represents “one or more”; cross-stroke arrowhead represents “exactly one”. Blue numbers indicate the number of variables extracted from each associated table for each visit. B, inclusion/exclusion procedure for the present study. Double-line arrows indicate the procession of visit records. C, Venn diagram of case cohort sizes for each of the five CDI case definitions before matching, including sizes of all intersections between case definitions (overlaps). Areas are not to scale. There is no intersection between case definitions (ii) and (iii), since only the first positive toxin assay result for each visit was examined. Case definition (iv), “by EIA or PCR (+),” is a strict superset of case definitions (ii) and (iii). Case definition (v), “by any of these,” is a strict superset of case definitions (i), (ii), and (iii). Sizes of matched case cohorts are provided in Table 1. EMR, electronic medical record; CDI, Clostridium difficile infection.
Study Population
The cohort included all patients 18 years of age or older admitted between January 1, 2009 and October 22, 2015 (Figure 1B). For each patient, visits following the first recorded visit in the time range were excluded so that each patient corresponded to a single visit. Visits involving a patient death, defined as a recorded time of death within 24 hours after discharge, were excluded (2,682 adult patients; 1.5%). Visits with missing or invalid date information were excluded (<0.01% of all records).
Study Design
Prior studies vary on the use of ICD-9 discharge codes vs. positive laboratory tests to define CDI cases5,6 and identify differing positive predictive values for immunoassay and nucleic acid based laboratory tests.25–27 To ensure maximally robust results and allow comparison with prior studies, we repeated our analysis for five definitions of CDI:
An “008.45” ICD-9 visit diagnosis code
≥1 positive stool toxin enzyme immunoassay (EIA) lab result
≥1 positive stool toxin polymerase chain reaction (PCR) lab result
Either ii or iii
Any of i, ii, or iii
Our study’s time range included both a period where the EIA assay was the standard hospital laboratory test (~3 years) followed by a period where the PCR assay was standard (~4 years). For case cohorts (ii) and (iii), comparisons were only permitted with controls from the time range during which that same test was standard. The hospital laboratory protocol requires unformed stool samples for either toxin assay.
Statistical Analysis
Details of propensity model development, matching, evaluation of matching performance, and LOS comparisons are available in Supplementary Methods. Briefly, propensity models for CDI based on the five case definitions were trained using logistic regression with elastic net regularization. After exact matching on gender and age bins, nearest-neighbor 1:1 matching on the propensity score was performed with a caliper of 0.2 standard deviations of the logit of the propensity score (Figure S1).28 Matching was repeated using the matched controls against remaining unmatched controls to create a matched-again cohort, testing whether matching alone associates with changes in LOS. For each case definition of CDI, differences of the median LOS between cases and matched controls were calculated, and statistical significance tested with the two-sided Mann-Whitney U. Although violation of the proportional hazards assumption (Figure S2) pre-empted traditional Cox survival analysis, non-parametric Kaplan-Meier estimates of the time-dependent risk of discharge were plotted for matched cohorts.
To further address the possible effect of time-to-infection on CDI risk and measured LOS differences, we repeated the analysis for case definition (iv) stratified by the time of the first positive toxin assay, using three ranges: 0–3 days, 3–8 days, and ≥8 days. Propensity models were again fitted to each of these case cohorts for matching as described previously, with the added condition that controls discharged before the start of the CDI time window were ineligible for matching.29 LOS comparisons followed the same procedure as above. We furthermore fit a nonparametric multistate model consistent with previous studies,7,24,30 under which the mean excess LOS was estimated as the average difference in LOS between patients that had or had not transitioned through the infected state for all timepoints, weighted by the distribution of times spent in the uninfected state.
Analyses were performed in R 3.2.2, and all software code is available at: https://github.com/powerpak/cdi-cost
Results
371,622 records of visits during the study time range were queried from the EMR, with 23,968 variables extracted for each visit (Figure 1A and 1B). After filtering for the index visit per adult patient and excluding deaths and invalid dates, 171,938 visits were eligible for inclusion and classified into five overlapping case definitions for CDI. Case cohort sizes before matching and their overlaps are depicted in Figure 1C. Regularized logistic regression models predicting the risk of CDI acquisition were fitted to EMR data from the first 24 hours of each admission for each case definition, with consistently high predictive performance (Supplementary Methods; Figure S3).
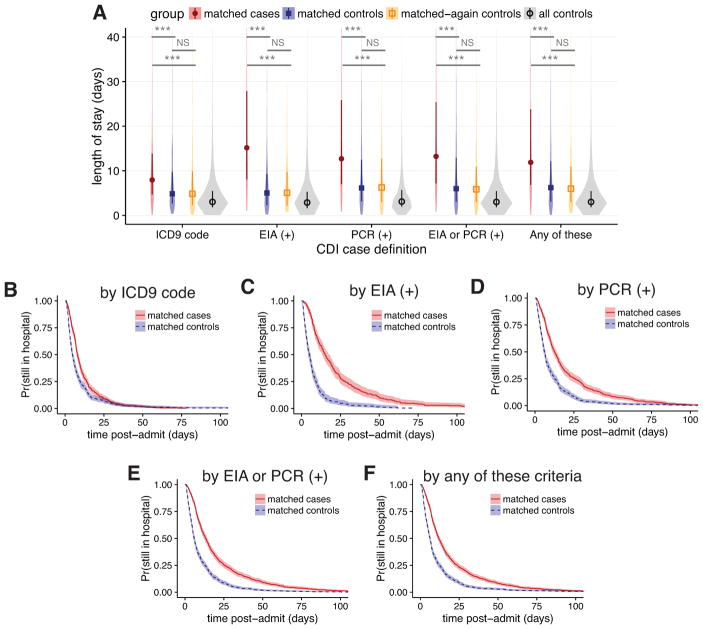
For each case definition, over 75% of cases were successfully matched by propensity score to controls (Figure 1C and Table 1). The groups are well matched on demographics and propensity scores (Table 1 and Figure S4). Differences in the median LOS between matched case and control cohorts for all CDI case definitions were strongly statistically significant, although the magnitude of the differences varied greatly between definitions (Figure 2A). The differences in the median LOS by case definition were: (i) by ICD-9 code, 3.1 days (95% confidence interval [CI], 2.2–3.9); (ii) by positive toxin EIA, 10.1 days (95% CI, 7.3–12.2), (iii) by positive toxin PCR, 6.6 days (95% CI, 5.0–8.1), (iv) by either toxin assay, 7.2 days (95% CI, 5.8–8.3); and (v) by any of these, 5.7 days (95% CI, 4.5–6.6). There were no significant differences in LOS for a second round of matching between matched controls and remaining controls (matched-again controls) for any of the case definitions (Figure 2A). Kaplan-Meier curves for the time-dependent risk of being discharged from the hospital showed significant differences between matched case and control cohorts up to post-admit day 60 for all case definitions except ICD-9 code (Figure 2B–F).
Table 1.
Demographic Characteristics of the Study Population and Matched Cohorts
| Characteristic | No. (%) | Matched cohorts for each CDI case definition | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| i. by ICD-9 code | ii. by EIA (+) | iii. by PCR (+) | iv. by EIA or PCR (+) | v. by any of the criteria | ||||||||||||
|
| ||||||||||||||||
| No. (%) | SMD after matching (P value) |
No. (%) | SMD after matching (P value) |
No. (%) | SMD after matching (P value) |
No. (%) | SMD after matching (P value) |
No. (%) | SMD after matching (P value) |
|||||||
|
|
|
|
|
|
||||||||||||
| All patients (n=171 936) |
All controls (n=171 356) |
Matched cases & controlsa (n=489) |
All controls (n= 73 647) |
Matched cases & controlsa (n=274) |
All controls (n=97 351) |
Matched cases & controlsa (n=493) |
All controls (n=170 994) |
Matched cases & controlsa (n=788) |
All controls (n=170 846) |
Matched cases & controlsa (n=945) |
||||||
| Female sex | 101 964 (59) | 101 638 (59) | 278 (57) | 0 (1) | 44 132 (60) | 145 (53) | 0 (1) | 57 340 (59) | 254 (52) | 0 (1) | 101 469 (59) | 408 (52) | 0 (1) | 101 390 (59) | 493 (52) | 0 (1) |
| Ageb | ||||||||||||||||
| 18–29 | 22 344 (13) | 22 266 (13) | 69 (14) | 0.016 (0.86) | 9 552 (13) | 22 (8) | 0.018 (0.79) | 12 714 (13) | 47 (10) | 0.005 (0.99) | 22 265 (13) | 79 (9) | 0.003 (0.98) | 22 245 (13) | 87 (9) | 0.004 (0.93) |
| 30–44 | 39 003 (23) | 38 898 (23) | 86 (18) | 16 451 (22) | 26 (9) | 22 430 (23) | 72 (15) | 38 879 (23) | 124 (13) | 38 845 (23) | 134 (14) | |||||
| 45–59 | 37 234 (22) | 37 129 (22) | 90 (18) | 15 956 (22) | 58 (21) | 21 069 (22) | 117 (24) | 37 025 (22) | 209 (23) | 36 999 (22) | 208 (22) | |||||
| 60–74 | 43 946 (26) | 43 802 (26) | 122 (25) | 18 407 (25) | 83 (30) | 25 273 (26) | 136 (28) | 43 680 (26) | 266 (29) | 43 643 (26) | 267 (28) | |||||
| 75–90 | 26 167 (15) | 26 041 (15) | 106 (22) | 11 817 (16) | 70 (26) | 14 120 (15) | 114 (23) | 25 936 (15) | 231 (24) | 25 912 (15) | 217 (23) | |||||
| ≥90 | 3 244 (2) | 3 220 (2) | 16 (3) | 1 464 (2) | 15 (5) | 1 745 (2) | 7 (1) | 3 209 (2) | 35 (3) | 3 202 (2) | 32 (3) | |||||
Abbreviation: CDI, Clostridium difficile infection; ICD-9, International Classification of Diseases Ninth Revision; EIA, enzyme immunoassay; PCR, polymerase chain reaction; SMD, standardized mean difference.
Separate columns are unnecessary because 1:1 exact matching was performed on the characteristics shown, and therefore all values are identical.
SMD is shown for age treated as a continuous variable; coarsened exact matching was performed using the listed age ranges.
Figure 2. Changes in length of stay for five case definitions of Clostridium difficile infection, not accounting for time of infection.
A, violin plots of the distributions in length of stay for matched cases, matched controls, matched-again controls, and all controls, for each of the five case definitions. Darker points and vertical bars depict the median and interquartile range for each group. Horizontal bars depict Mann-Whitney U tests for significance of differences between groups (***, Bonferroni-corrected P < 0.001; NS, not significant [P > 0.1]). B–F, Kaplan-Meier plots of the time-dependent probability for a patient to still be in the hospital, comparing matched cases and controls for each case definition of CDI. Shaded areas depict 95% confidence intervals calculated from standard errors. CDI, Clostridium difficile infection; ICD9, International Classification of Diseases Ninth Revision; EIA, enzyme immunoassay; PCR, polymerase chain reaction.
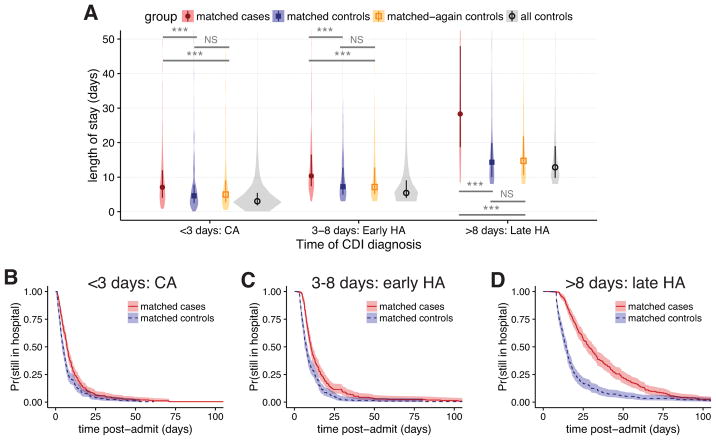
Estimates of LOS associated with CDI are inflated by dependencies on time-to-infection—if longer pre-infection LOS increases CDI risk, i.e., reverse causation, this leads to overestimates in attributable cost.7,24 We therefore performed two follow-up analyses to account for this. First, we stratified the LOS comparison by the time of CDI diagnosis for case definition (iv) into 0–3 day, 3–8 day, and ≥8 day case cohorts, training new propensity models for re-matching, with similar performance (Figure S5). Since 3 days is a typical cutoff for differentiating community acquired (CA) from healthcare-associated (HA) CDI,25,31 these strata were named “CA,” “early HA,” and “late HA,” respectively. As suspected, stratification revealed a positive correlation between time of diagnosis and CDI-associated difference in LOS (Figure 3A). The differences in medians were: for CA, 2.5 days (95% CI, 1.2–3.4); early HA, 3.1 days (95% CI, 1.8–4.4); and late HA, 14.0 days (95% CI, 9.9–17.1). All comparisons between matched cases and controls were again strongly statistically significant, and comparisons with again-matched controls were not significant (Figure 3A). Kaplan-Meier plots likewise confirmed a correlation between time of CDI diagnosis and differences in time-dependent discharge risk (Figure 3B–D).
Figure 3. Changes in length of stay for Clostridium difficile infection defined by any positive toxin assay, stratified by the time to infection.
A, violin plots of the distributions in length of stay for matched cases, matched controls, matched-again controls, and all controls, for three ranges of the result time for the first positive toxin assay. Points and vertical bars depict the median and interquartile range for each group. Horizontal bars depict Mann-Whitney U tests for significance of differences between groups (***, Bonferroni-corrected P < 0.001; NS, not significant [P > 0.1]). B–D, Kaplan-Meier plots of the time-dependent probability for a patient to still be in the hospital, comparing matched cases and controls for the same three ranges of the time of the first positive toxin assay. Shaded areas depict 95% confidence intervals calculated from standard errors. CDI, Clostridium difficile infection; CA, community acquired; HA, healthcare associated.
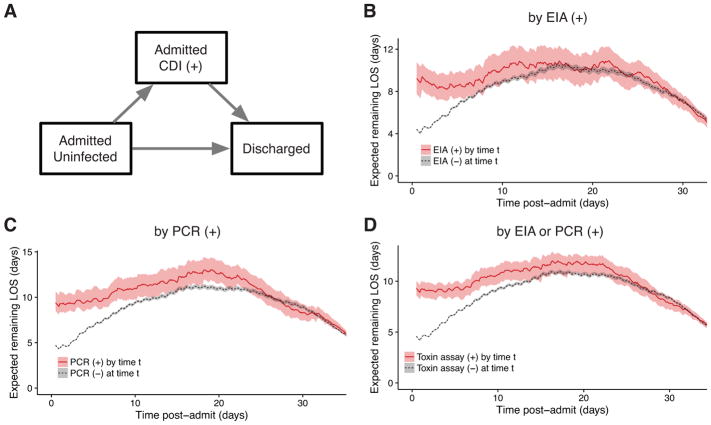
To further address reverse causation, we fit a multistate model similar to previously published studies7,24,30 that explicitly estimates time-dependent, competing risks of transitioning to CDI vs. discharge. Figure 4A depicts the model’s states and transitions. After fitting the model for the case definitions with a time of diagnosis (ii, iii, and iv), the expected remaining LOS can be compared across cohorts that have already transitioned to the CDI infected state vs. those that are still CDI negative at any given timepoint (Figure 4B–D). To summarize the overall relationship between CDI and LOS, differences in LOS were weighted by the distribution of times spent in the initial state and averaged. The average differences for each case definition were: (ii) by positive toxin EIA, 3.0 days (95% CI, 2.0–4.0); (iii) by positive toxin PCR, 3.5 days (95% CI, 2.7–4.5); and (iv) by either toxin assay, 3.3 days (95% CI, 2.6–4.0). Notably, the 95% CI for the difference in cohort (iv) overlaps the 3.1 day difference for the “early HA” stratum of the propensity-matched analysis in the same cohort.
Figure 4. Multistate model of expected remaining length of stay for Clostridium difficile infection case definitions involving toxin assays.
A, three states of the multistate model and allowed transitions. Patients may only transition in the direction of the arrows. B–D, expected remaining LOS for each post-admit time t depending on whether the patient has had a positive (+) toxin assay by that timepoint, for each of the case definitions involving toxin assays. Shaded areas depict 95% confidence intervals calculated from 1,000 bootstrap samples. CDI, Clostridium difficile infection; EIA, enzyme immunoassay; PCR, polymerase chain reaction; LOS, length of stay.
Discussion
This study examined nearly seven years of uncurated EMR data for a single hospital and determined associated costs of CDI as defined by either visit diagnosis codes or lab results. In the analysis unadjusted for time-to-infection, differences in LOS were often greater than national averages from similar unadjusted studies,3,5,6 but changes in the case definition resulted in substantial changes in the estimated differences in LOS. Although two hospitals reported good concordance between ICD-9 codes and CDI toxin assay results,32,33 this is not necessarily the case for all hospitals. We found that 75% of ICD-9 coded visits involved a positive toxin assay, while only 46% of visits with a positive toxin assay had the ICD-9 code (Figure 1C). Changes in LOS were not significantly different between EIA and PCR toxin assays, although our study was limited by a smaller sample size for EIA (+) cases. Toxin assays are likely a more reliable CDI definition given their basis in clinical symptoms and evidence for CDI, whereas medical coding suffers from biases introduced by billing and reimbursement.34,35
Treating CDI as a baseline condition by ignoring the relationship between pre-infection hospital exposure and CDI risk overestimates associated costs.7,24,36 Unlike visit diagnosis codes, toxin assay results provide a presumptive time-to-infection that we incorporated into two different statistical methods addressing time-dependent bias. When using a case definition of either toxin assay being positive, the measured difference in LOS in the multistate model corresponded closely with the difference seen in the “early HA” stratum of a time-stratified propensity-matched analysis (3.3 vs. 3.1 days). This suggests that measured differences in this study robustly reflect associated costs of HA-CDI in our patient population. Since estimates for each time-to-infection stratum in the matching analysis differed greatly (Figure 3), time-to-infection clearly contributed bias to the unstratified analysis (Figure 2), demonstrating how the many studies that ignore this bias3,5,6 produce inflated estimates. In our dataset, ignoring time-dependent bias would lead to a more than two-fold overestimation of CDI-associated LOS. Given our findings, we cautiously interpret the results of meta-analyses that conflate ICD-9 code and toxin assay case definitions and often ignore time-dependent bias.4–6
To our knowledge, this is the first study that uses machine learning on uncurated EMR data to estimate the local cost of CDI. Our models of CDI risk performed on par with prior models fitted to lower-dimensional data.23,37,38 Since our models are based on tens of thousands of structured fields in the EMR that require neither chart review nor manual curation beyond masking known CDI-related effects, re-analysis of future data is inexpensive. Starting from exported visit data, the entire analysis runs in several hours on standard desktop computers. Therefore, the effects of new interventions against CDI can be efficiently monitored over time, e.g., continually testing whether new treatments actually lower the CDI-associated LOS or quantifying cost savings of new preventive strategies that decrease CDI incidence. Changes in LOS can be extrapolated to approximate economic costs by multiplying by the average cost of extra inpatient-days as LOS is the main contributor to CDI’s cost.3,21,22,36 In our dataset, using the time-dependency adjusted differences in LOS of 3.1–3.3 days and the national average cost of additional inpatient-days for CDI cases,3 the median cost associated with each case would be approximately $10,600–11,300; this is substantial in comparison to the national average price for an inpatient visit—approximately $13,000 in 2011.11 Using the average yearly caseload observed in the dataset for toxin assay positive cases, our figures represent an annual accounting cost to Mount Sinai of approximately $1.5 million, not including the opportunity cost of bed occupancy by CDI patients or the impact on infection control resources.36 In principle, our analysis is generalizable to any HAI where lab results recorded in the EMR robustly reflect the incidence of infections.
Our study has several limitations. The analysis was designed conservatively, preferring that models underestimate rather than overestimate CDI-associated changes. For example, we censored all patient visits ending in death; therefore, our results are conditioned on patient survival, although a sensitivity analysis that included 12–16% additional cases ending in patient death yielded similar quantitative and qualitative results. Additionally, restricting to one index visit per patient certainly excluded many repeat visits for recurrent CDI, which are known to incur higher costs.12,13,39 We preferred a relatively simple, fast machine learning technique, elastic net regularized generalized linear models, whereas more advanced techniques might marginally improve propensity model accuracy.
Propensity score matching itself has been criticized for potentially introducing bias via collider variables.40 However, substantial empirical comparisons of estimates from observational and randomized controlled trial data show that propensity matching often reduces bias.41 Recent investigations of penalized regression propensity matching also show a reduction in bias.42,43 We believe our implementation reduced bias, as our estimate of the effect of CDI on LOS demonstrated significant deviations from unmatched analyses and concordance with the multistate matching analysis (which did not leverage propensity scores or matching). We also note again that propensity matched estimates offer a conservative effect size, which was the intention of this study.
EMR data has known drawbacks compared to clinical research data, such as limitations in time precision, the sparsity of the data, and increased opportunity for coding error. We did not have structured billing data, so we cannot characterize the exact relationship between LOS and costs beyond the proportional estimate above. Finally, only one hospital’s data was available for this study. We provide complete code for our analysis so that it may be re-implemented elsewhere and improved by the community.
In conclusion, two independent statistical analyses adjusting for time-dependent bias produced similar results for the CDI-associated change in LOS at Mount Sinai (3.1 and 3.3 days), suggesting that automated methods based on machine learning and uncurated EMR data robustly and conservatively estimate the local cost of an HAI in both LOS and financial terms. This procedure is transparent, reproducible, and inexpensive, suggesting that hospitalists and infection control officers can leverage EMR data to estimate their specific, local costs of HAIs on an ongoing basis rather than relying on widely varying benchmarks published by other institutions.
Supplementary Material
Acknowledgments
We thank Deena Altman, Camille Hamula, and Gopi Patel for their assistance in improving the design of the study and reviewing the manuscript.
Financial support
This work was supported by the Icahn Institute for Genomics and Multiscale Biology at Mount Sinai, and also in part by NIAID (F30AI122673 and R01AI119145), and the resources and expertise of the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Footnotes
Potential conflicts of interest
ERS receives salary support from and acts as an advisor for Sema4 Incorporated. All other authors report no conflicts of interest relevant to this article.
References
- 1.Leffler DA, Lamont JT. Clostridium difficile. N Engl J Med. 2015;372:1539–48. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 2.Davies KA, Longshaw CM, Davis GL, et al. Underdiagnosis of Clostridium difficile across Europe: The European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID) Lancet Infect Dis. 2014;14(12):1208–1219. doi: 10.1016/S1473-3099(14)70991-0. [DOI] [PubMed] [Google Scholar]
- 3.Zimlichman E, Henderson D, Tamir O, et al. Health care-associated infections: a meta-analysis of costs and financial impact on the US health care system. JAMA Intern Med. 2013;173(22):2039–46. doi: 10.1001/jamainternmed.2013.9763. [DOI] [PubMed] [Google Scholar]
- 4.Ghantoji SS, Sail K, Lairson DR, DuPont HL, Garey KW. Economic healthcare costs of Clostridium difficile infection: A systematic review. J Hosp Infect. 2010;74(4):309–318. doi: 10.1016/j.jhin.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Palazuelos-Munoz S, Balsells EM, Nair H, Chit A, Kyaw MH. Cost of hospital management of Clostridium difficile infection in United States—a meta-analysis and modelling study. BMC Infect Dis. 2016;16(1):447. doi: 10.1186/s12879-016-1786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabriel L, Beriot-Mathiot A. Hospitalization stay and costs attributable to Clostridium difficile infection: A critical review. J Hosp Infect. 2014;88(1):12–21. doi: 10.1016/j.jhin.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Stevens VW, Khader K, Nelson RE, et al. Excess Length of Stay Attributable to Clostridium difficile Infection (CDI) in the Acute Care Setting: A Multistate Model. Infect Control Hosp Epidemiol. 2015;36(Cdi):1–7. doi: 10.1017/ice.2015.132. [DOI] [PubMed] [Google Scholar]
- 8.Lofgren ET, Cole SR, Weber DJ, Anderson DJ, Moehring RW. Hospital-Acquired Clostridium difficile Infections: Estimating All-Cause Mortality and Length of Stay. Epidemiology. 2014;25(4):570–575. doi: 10.1097/EDE.0000000000000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz MH. Pay for preventing (not causing) health care-associated infections. JAMA Intern Med. 2013;173(22):2046. doi: 10.1001/jamainternmed.2013.9754. [DOI] [PubMed] [Google Scholar]
- 10.Dubberke ER, Carling P, Carrico R, et al. Strategies to Prevent Clostridium difficile Infections in Acute Care Hospitals: 2014 Update. Infect Control Hosp Epidemiol. 2014;35(6):628–645. doi: 10.1086/676023. [DOI] [PubMed] [Google Scholar]
- 11.Cooper Z, Craig S, Gaynor M, Van Reenen J. The Price Ain’t Right? Hospital Prices and Health Spending on the Privately Insured. NBER Work Pap. 2015:21815. doi: 10.3386/w21815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubberke ER, Schaefer E, Reske KA, Zilberberg M, Hollenbeak CS, Olsen MA. Attributable inpatient costs of recurrent Clostridium difficile infections. Infect Control Hosp Epidemiol. 2014;35(11):1400–1407. doi: 10.1086/678428. [DOI] [PubMed] [Google Scholar]
- 13.Dubberke ER, Reske KA, Olsen MA, McDonald LC, Fraser VJ. Short- and Long-Term Attributable Costs of Clostridium difficile-Associated Disease in Nonsurgical Inpatients. Clin Infect Dis. 2008;46(4):497–504. doi: 10.1086/526530. [DOI] [PubMed] [Google Scholar]
- 14.Greco G, Shi W, Michler RE, et al. Costs associated with health care-associated infections in cardiac surgery. J Am Coll Cardiol. 2015;65(1):15–23. doi: 10.1016/j.jacc.2014.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry J, Pylypchuk Y, Searcy T, Patel V. Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals: 2008–2015. ONC Data Br. 2016;(35) Available at: https://dashboard.healthit.gov/evaluations/data-briefs/non-federal-acute-care-hospital-ehr-adoption-2008-2015.php.
- 16.Gray BH, Bowden T, Johansen I, Koch S. Electronic health records: an international perspective on “meaningful use”. Issue Brief (Commonw Fund) 2011 Nov;28:1–18. [PubMed] [Google Scholar]
- 17.Etheredge LM. A rapid-learning health system. Health Aff (Millwood) 2007;26(2):w107–18. doi: 10.1377/hlthaff.26.2.w107. [DOI] [PubMed] [Google Scholar]
- 18.Dahabreh IJ, Kent DM. Can the learning health care system be educated with observational data? JAMA. 2014;312(2):129–30. doi: 10.1001/jama.2014.4364. [DOI] [PubMed] [Google Scholar]
- 19.Pak TR, Kasarskis A. How Next-Generation Sequencing and Multiscale Data Analysis Will Transform Infectious Disease Management. Clin Infect Dis. 2015;61(11):1695–1702. doi: 10.1093/cid/civ670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krumholz HM, Terry SF, Waldstreicher J. Data Acquisition, Curation, and Use for a Continuously Learning Health System. JAMA. 2016;316(16):1669. doi: 10.1001/jama.2016.12537. [DOI] [PubMed] [Google Scholar]
- 21.Wilcox MH, Cunniffe JG, Trundle C, Redpath C. Financial burden of hospital-acquired Clostridium difficile infection. J Hosp Infect. 1996;34(1):23–30. doi: 10.1016/S0195-6701(96)90122-X. [DOI] [PubMed] [Google Scholar]
- 22.McGlone SM, Bailey RR, Zimmer SM, et al. The economic burden of Clostridium difficile. Clin Microbiol Infect. 2012;18(3):282–289. doi: 10.1111/j.1469-0691.2011.03571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiens J, Campbell W. Learning Data-Driven Patient Risk Stratification Models for Clostridium difficile. Open Forum Infect Dis. 2014;1(2):1–9. doi: 10.1093/ofid/ofu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell BG, Gardner A, Barnett AG, Hiller JE, Graves N. The prolongation of length of stay because of Clostridium difficile infection. Am J Infect Control. 2014;42(2):164–167. doi: 10.1016/j.ajic.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Polage CR, Gyorke CE, Kennedy MA, et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med. 2015;175(11):1–10. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagdasarian N, Rao K, Malani PN. Diagnosis and Treatment of Clostridium difficile in Adults. JAMA. 2015;313(4):398. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moehring RW, Lofgren ET, Anderson DJ. Impact of Change to Molecular Testing for Clostridium difficile Infection on Healthcare Facility-Associated Incidence Rates. Infect Control Hosp Epidemiol. 2013;34(10):1055–61. doi: 10.1086/673144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Austin PC. Optimal caliper widths for propensity-score matching when estimating differences in means and differences in proportions in observational studies. Pharm Stat. 2011;10(2):150–161. doi: 10.1002/pst.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YP, Propert KJ, Rosenbaum PR. Balanced Risk Set Matching. J Am Stat Assoc. 2001 Oct;96:870–882. doi: 10.1198/016214501753208573. 2014. [DOI] [Google Scholar]
- 30.van Kleef E, Green N, Goldenberg SD, et al. Excess length of stay and mortality due to Clostridium difficile infection: A multi-state modelling approach. J Hosp Infect. 2014;88(4):213–217. doi: 10.1016/j.jhin.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Longtin Y, Paquet-Bolduc B, Gilca R, et al. Effect of Detecting and Isolating Clostridium difficile Carriers at Hospital Admission on the Incidence of C difficile Infections: A Quasi-Experimental Controlled Study. JAMA Intern Med. 2016;176(6):796–804. doi: 10.1001/jamainternmed.2016.0177. [DOI] [PubMed] [Google Scholar]
- 32.Dubberke ER, Reske KA, McDonald LC, Fraser VJ. ICD-9 codes and surveillance for Clostridium difficile-associated disease. Emerg Infect Dis. 2006;12(10):1576–1579. doi: 10.3201/eid1210.060016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheurer DB, Hicks LS, Cook EF, Schnipper JL. Accuracy of ICD-9 coding for Clostridium difficile infections: a retrospective cohort. Epidemiol Infect. 2007;135(6):1010–3. doi: 10.1017/S0950268806007655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee C, Murphy MV, Li L, Platt R, Klompas M. Improving documentation and coding for acute organ dysfunction biases estimates of changing sepsis severity and burden: a retrospective study. Crit Care. 2015;19(1):1–11. doi: 10.1186/s13054-015-1048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romano PS, Mark DH. Bias in the coding of hospital discharge data and its implications for quality assessment. Med Care. 1994;32(1):81–90. doi: 10.1097/00005650-199401000-00006. [DOI] [PubMed] [Google Scholar]
- 36.Graves N, Harbarth S, Beyersmann J, Barnett A, Halton K, Cooper B. Estimating the cost of health care-associated infections: mind your p’s and q’s. Clin Infect Dis. 2010;50(7):1017–1021. doi: 10.1086/651110. [DOI] [PubMed] [Google Scholar]
- 37.Dubberke ER, Yan Y, Reske KA, et al. Development and validation of a Clostridium difficile infection risk prediction model. Infect Control Hosp Epidemiol. 2011;32(4):360–6. doi: 10.1086/658944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanner J, Khan D, Anthony D, Paton J. Waterlow score to predict patients at risk of developing Clostridium difficile-associated disease. J Hosp Infect. 2009;71(3):239–244. doi: 10.1016/j.jhin.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigues R, Barber GE, Ananthakrishnan AN. A Comprehensive Study of Costs Associated With Recurrent Clostridium difficile Infection. Infect Control Hosp Epidemiol. 2016:1–7. doi: 10.1017/ice.2016.246. [DOI] [PubMed] [Google Scholar]
- 40.Pearl J. Technical Report R–348. University of California; Los Angeles, CA: 2009. Myth, Confusion, and Science in Causal Analysis. Available at: http://ftp.cs.ucla.edu/pub/stat_ser/r348.pdf. [Google Scholar]
- 41.Lonjon G, Boutron I, Trinquart L, et al. Comparison of treatment effect estimates from prospective nonrandomized studies with propensity score analysis and randomized controlled trials of surgical procedures. Ann Surg. 2014;259(1):18–25. doi: 10.1097/SLA.0000000000000256. [DOI] [PubMed] [Google Scholar]
- 42.Athey S, Imbens GW, Wager S. Approximate Residual Balancing: De-Biased Inference of Average Treatment Effects in High Dimensions. arXiv. 2016 Available at: http://arxiv.org/abs/1604.07125.
- 43.Antonelli J, Cefalu M, Palmer N, Agniel D. Double robust matching estimators for high dimensional confounding adjustment. arXiv. 2016 doi: 10.1111/biom.12887. Available at: http://arxiv.org/abs/1612.00424. [DOI] [PMC free article] [PubMed]
- 44.Halpin T, Morgan T. Information Modeling and Relational Databases. 2. Elsevier Science; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.