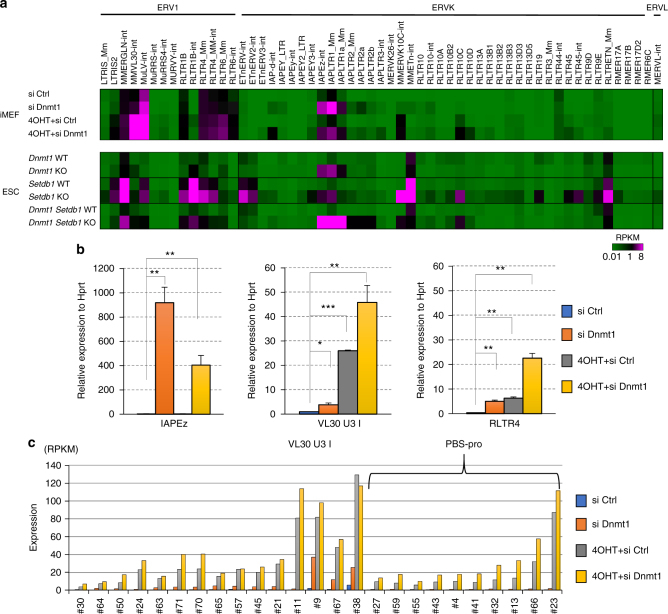
Fig. 4.
Dnmt1 and Setdb1 are required for the silencing of distinct sets of ERVs. a Expression of ERV families by the loss of Dnmt1, Setdb1, and simultaneous depletion of Dnmt1 and Setdb1 in iMEFs and ESCs. To obtain mRNA-seq data of iMEFs, Setdb1 cKO iMEFs were transfected with siRNA against Dnmt1 or control siRNA in combination with or without Setdb1 depletion by the 4OHT treatment, then mRNA was isolated. RNA-seq data from Dnmt1 cKO or double cKO of Dnmt1 and Setdb1 ESCs (6 days after treatment of 4OHT or without treatment)36 were reanalyzed. Heatmap indicates the magnitude of the RPKM value. b Setdb1 cKO iMEFs were transfected with siRNA against Dnmt1 either alone or in combination with the loss of Setdb1. RT-qPCR of VL30 U3 class I RLTR6, IAPEz, and RLTR4 was performed. Values represent mean expression relative to VL30 U3 I control (n = 2 biological replicates). Error bars represent s.d. *0.005 < P < 0.05, **0.0005 < P < 0.005, ***P < 0.0005, Student’s t-test. c The derepression induced by Dnmt1 KD, Setdb1 KO, and simultaneous depletion of Dnmt1 and Setdb1 in iMEFs in each VL30 U3 class I element (totally 71 elements) is shown. The number of RNA-seq reads overlapping with a locus was divided by the length of the locus and normalized by million mapped reads (RPKM) as shown in Fig. 3c top