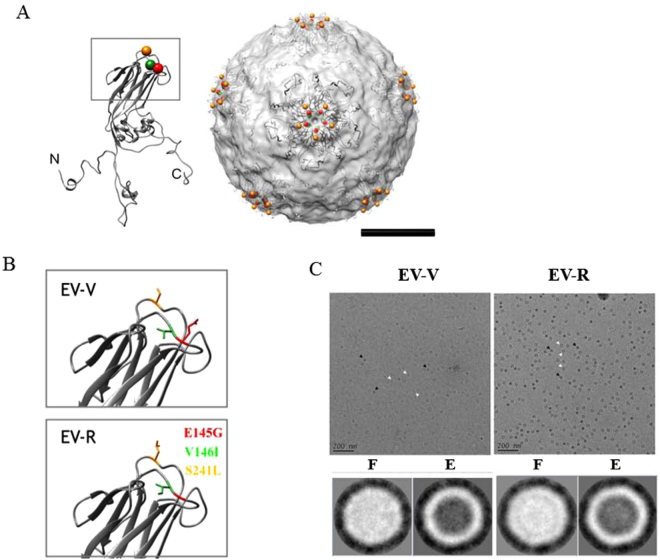
Figure 6.
Modelling of the steric structure of EV71 particles and EV71 mutants. EV-V and EV-R particles were prepared for TEM imaging as described in the Methods. (A) The atomic structure of VP1 extracted from VP1-4 complex (PDB: 3VBS) was fitted into cryoEM structure of EV71 (EMDB: 5558). The spheres showed the residues 145 (colored in red), 146 (colored in green), and 241 (colored in orange) surrounding the 5-fold axis of the structure. The N, C terminus are indicated. The scale bar = 10 nm. (B) The 3-D configurations of the side chains of the residues at VP1-145, 146, and 241 from EV-V and EV-R are shown. The VP1 of EV-R was modelled from SWISSMODE website, a protein structure homology-modelling server. The residues were colored as previous figure. (C) The cryoEM images (upper panel) and 2-D classification (lower panel) of EV71 particles derived from Vero cell line (EV-V, left panel) and from RD cell lone (EV-R, right panel). The image analyses showed that the empty (E) and full (F) particles indicated by white and black arrows, respectively existed in the samples and the diameter of EV71 particles were approximately 30 nm. The scale bar in the micrography is 200 nm. The particle picking and 2-D classification analyses were done by EMAN2 and Xmipp where the contrast was inverted.