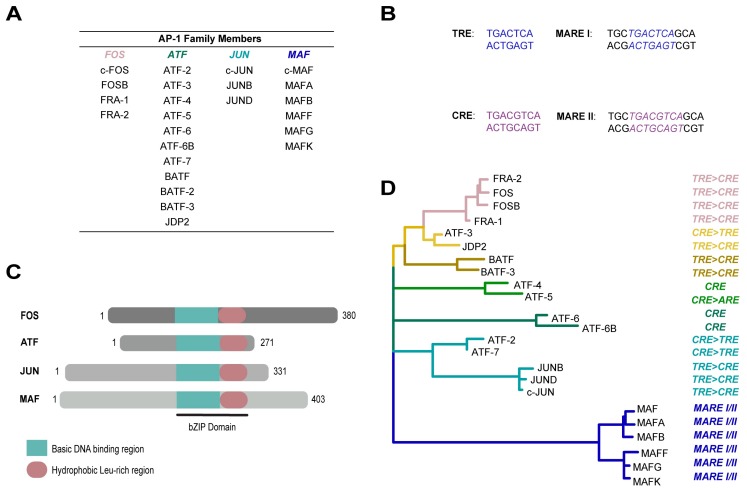
Figure 1.
The AP-1 transcription factor family members. (A) Table of the different AP-1 transcription factor family subgroups FOS, ATF, JUN and MAF; (B) The different DNA response-elements (REs) that AP-1 TFs recognise and to which they bind. The 12-O-tetradecanoylphorbol-13-acetate response element (TRE) is the most common; however, depending on dimer configuration, AP-1 TFs can bind to additional elements, such as CRE, MARE I, and MARE II. Note MARE I is an extension of TRE, whereas MARE II is an extension of CRE; (C) Schematic representation of the structure of AP-1 proteins including FOS, ATF, JUN, and MAF. AP-1 TFs share two common regions, the basic motif and the leucine zipper; together these regions form the bZIP domain. Sequence data was exported from Nextprot; (D) Phylogenetic tree of AP-1 transcription factor family members and their binding REs. The amino acid sequences were aligned with ClustalW implemented with MEGA 7.0.21. The alignment was manually rearranged in Bioedit 7.0.8.0. In order to find the best-fit amino acid substitution model for the alignment, a model test was performed with MEGA 7.0.21. The Maximum Likelihood tree was also calculated with MEGA 7.0.21, applying the parameters obtained from Modeltest.