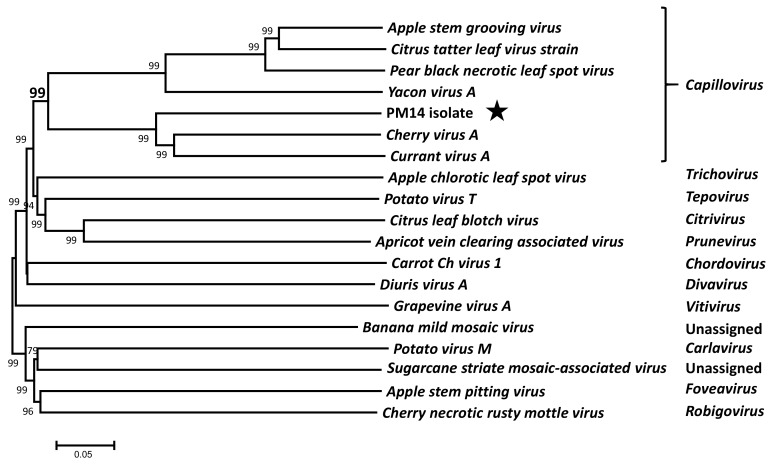
Figure 2.
Unrooted phylogenetic tree reconstructed using the complete genome sequences of representative Betaflexiviridae members. The tree was constructed using the neighbor-joining method, and the statistical significance of branches was evaluated by bootstrap analysis (1000 replicates). Bootstrap values above 70% are shown. The scale bar represents 5% divergence between sequences. Sequences retrieved from GenBank are NC001749 Apple stem grooving virus; AY646511 Citrus tatter leaf virus; AY596172 Pear black necrotic leaf spot virus; NC030657 Yacon virus A; NC003689 Cherry virus A; NC029301 Currant virus A; NC001409 Apple chlorotic leaf spot virus; NC011062 Potato virus T; NC003877 Citrus leaf blotch virus; NC023295 Apricot vein clearing associated virus; NC025469 Carrot Ch virus 1; NC019029 Diuris virus A; NC003604 Grapevine virus A; NC002729 Banana mild mosaic virus; NC001361 Potato virus M; NC003870 Sugarcane striate mosaic-associated virus; and NC002468 Cherry necrotic rusty mottle virus. The genus to which each virus belongs is indicated at the right. The PM14 isolate is indicated by a black star.