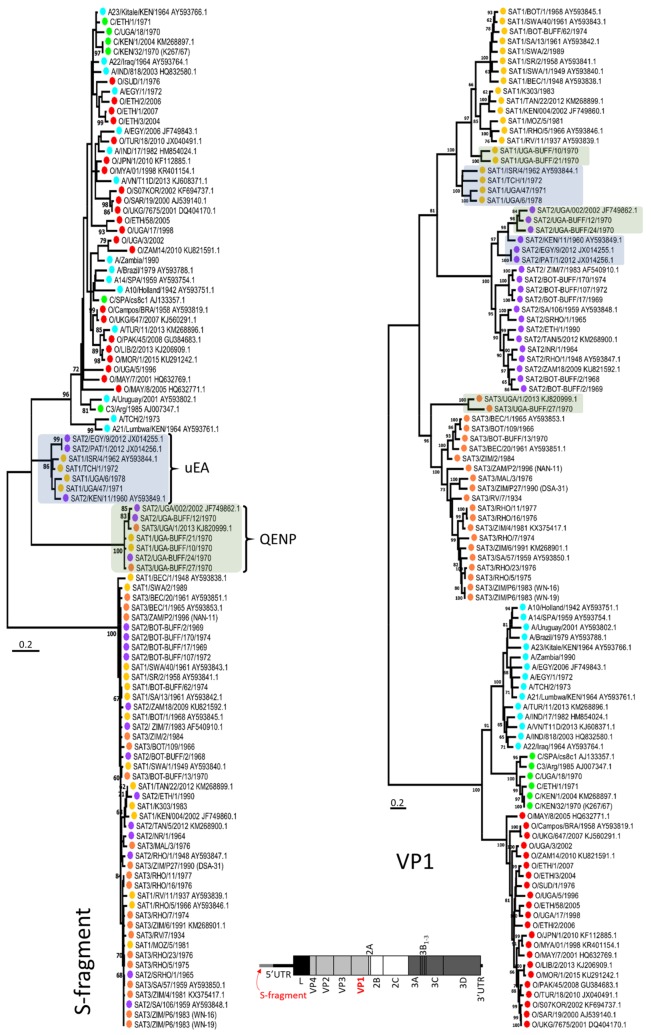
Figure 1.
Maximum likelihood (ML) phylogenetic analysis of S-fragment sequences defining two new viral genetic clades (isolates from the Queen Elizabeth National Park (QENP) and “unusual” sequences from East Africa (uEA)). The evolutionary history based on S-fragment and VP1 sequence was inferred using the ML method and general time reversible nucleotide substitution model with gamma distribution and invariant sites (GTR + G + I) (both implemented in MEGA7 software). For each fragment the tree with the highest log likelihood is shown. The percentage of trees (with the 60% cut-off) in which the associated taxa clustered together is shown next to the branches. Initial tree for the heuristic search were obtained automatically by applying Neighbor-Joining (NJ) and BioNJ methods. Tip taxa were colour-coded according to their serotype: A—blue, C—green, O—red, Southern African Territories (SAT) 1—yellow, SAT 2—purple and SAT 3—orange. Isolates which (on the S-fragment tree) clustered in QENP and uEA clades were highlighted in green and blue, respectively. A schematic drawing of FMDV genome with highlighted in red S-fragment and VP1 encoding region is also shown. Taxa names contain full year date.