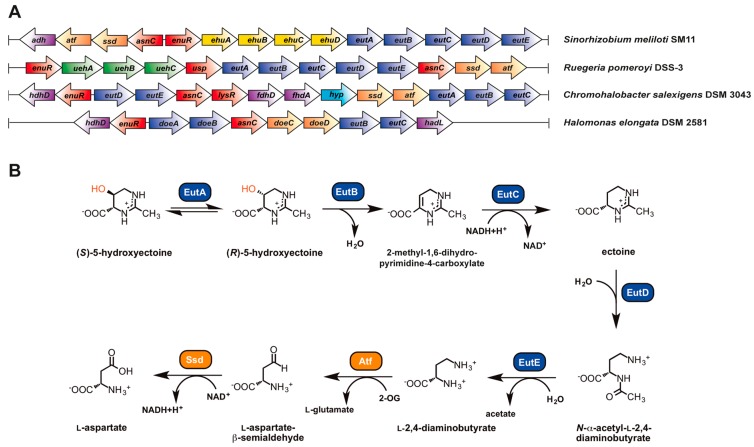
Figure 6.
Genetics and catabolic pathways for the utilization of ectoine and 5-hydroxyectoine as nutrients. (A) Genetic organization of the ectoine/5-hydroxyectoine-catabolic gene cluster in Sinorhizobium meliloti SM11 [265], Ruegeria pomeroyi DSS-3 [263,270], Halomonas elongata DSM 258 [158] and Chromohalobacter salexigens DSM 3043 (predicted from the genome sequence) [274]. In addition to the transporter and catabolic genes discussed in the main text, some of these gene clusters contain genes with yet undefined roles in ectoine catabolism. Their gene products have bioinformatically predicted functions as alcohol dehydrogenase (adh), hydroxyacid dehydrogenase (hdhD), formate dehydrogenases (fdhD, fdhA), haloacid dehalogenase (hadL), transcriptional regulator (lysR) and a hypothetical protein (hyp). (B) Predicted pathway for the catabolism of ectoine and its derivative 5-hydroxyectoine in R. pomeroyi DSS-3. The EutABC-enzymes are predicted to convert 5-hydroxyectoine in a three-step reaction into ectoine. The ectoine ring is subsequently hydrolyzed by the EutD enzyme, resulting in the production of N-α-ADABA, an intermediate, which is then further catabolized to l-aspartate by the EutE, Atf and Ssd enzymes. These data were compiled from the literature [158,263,270]. The ectoine-derived metabolites N-α-ADABA and l-2,4-diaminobutyrate (DABA) serve as inducers for the transcriptional control of the ectoine/5-hydroxyectoine import and catabolic gene clusters by the EnuR regulatory protein [270].