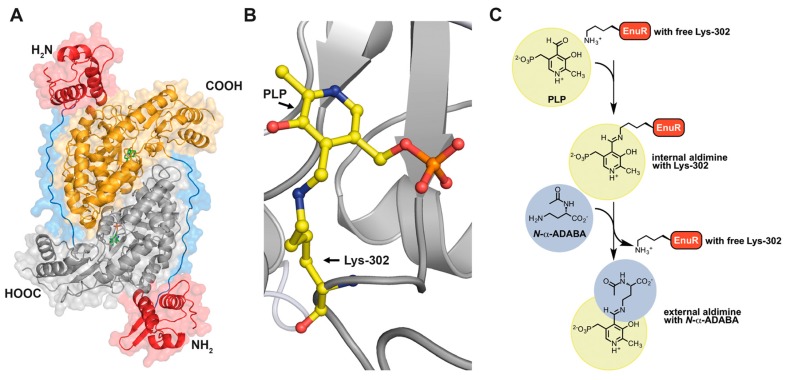
Figure 7.
EnuR, a PLP-containing transcriptional regulator of ectoine/5-hydroxyectoine gene clusters. (A) in silico model of the predicted Ruegeria pomeroyi DSS-3 EnuR dimer that was derived from the crystallographic structure of the Bacillus subtilis GabR (PDB accession code 4N0B) [279]. The EnuR model was built with the SWISS-MODEL web server (https://swissmodel.expasy.org/) [286] and visualized using the PyMOL Molecular Graphics System suit (https://pymol.org/2/) [287]. The C-terminal aminotransferase-domains of the EnuR dimer are shown in grey/yellow, the N-terminal DNA-binding domains are represented in red and the flexible linkers connecting these domains are depicted in blue. Each monomer contains a PLP molecule covalently bound via an Schiff base to Lys302 in the aminotransferase domain [263,270]. This internal aldimine [173] is depicted in (B) in a close-up view. (C) Model for the chemistry underlying binding and release of the inducer N-α-ADABA to the PLP-cofactor bound to Lys302 of the EnuR regulator. In the first step, PLP is covalently bound by the side-chain of Lys302 and thus forms an internal aldimine [173]. Upon binding of the inducer N-α-ADABA to PLP, PLP is released from Lys302 and an external aldimine [173] is formed. This sequence of events is envisioned to trigger a conformational change in EnuR, thereby altering its DNA-binding properties. This scheme for inducer binding by EnuR is based upon detailed biochemical and structural analysis of the B. subtilis GabR regulator that uses GABA as its inducer [279,280,281,282,283].