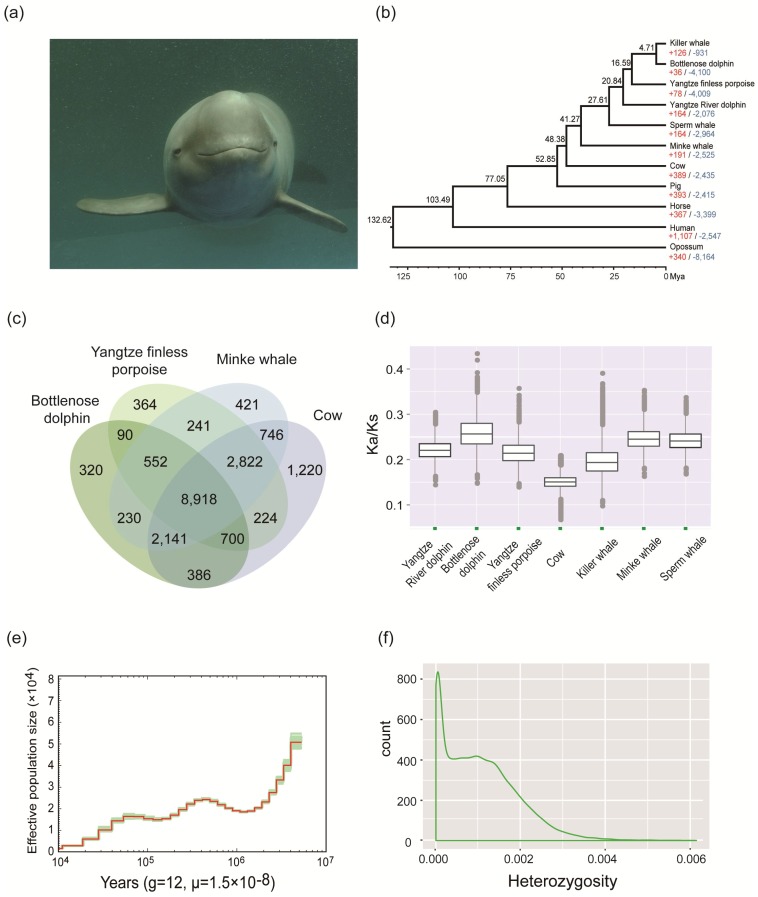
Figure 1.
Gene families, phylogenetic relationships, and demographic history of the Yangtze finless porpoise. (a) Picture of a Yangtze finless porpoise (image from SL); (b) Phylogenetic tree constructed using the maximum likelihood approach and a comparison of gene family numbers. Black numbers next to the branches indicate divergence times, while the red and blue numbers indicate the number of gene families that have expanded or contracted, respectively, since the split from the common ancestor; (c) Venn diagram showing unique and overlapping gene families in the Yangtze finless porpoise, common minke whale, bottlenose dolphin, and cow genomes. Each number represents a gene family number; (d) Box-plot showing ratios of non-synonymous to synonymous mutations (Ka/Ks) in the Yangtze finless porpoise, Yangtze River dolphin, bottlenose dolphin, cow, killer whale, common minke whale, and sperm whale genomes; (e) Demographic history of the Yangtze finless porpoise constructed using the pairwise sequentially Markovian coalescence model; (f) Distribution of heterozygosity in the Yangtze finless porpoise genome (heterozygosity ratios of non-overlapping 50 K windows).