Abstract
The LIM domain only protein LMO3 is a transcriptional regulator that has been shown to regulate several behavioral responses to alcohol. Specifically, Lmo3 null (Lmo3Z) mice consume more ethanol in a binge-drinking test and show enhanced ethanol-induced sedation. Due to the high comorbidity of alcohol use and anxiety, we investigated anxiety-like behavior in Lmo3Z mice. Lmo3Z mice spent more time in the open arms of the elevated plus maze compared with their wild-type littermates, but the effect was confounded by reduced locomotor activity. To verify the anxiety phenotype in the Lmo3Z mice, we tested them for novelty-induced hypophagia and found that they also showed reduced anxiety in this test. We next explored the mechanism by which LMO3 might regulate anxiety by measuring mRNA and protein levels of corticotropin releasing factor (encoded by the Crh gene) and its receptor type 1 (Crhr1) in Lmo3Z mice. Reduced Crhr1 mRNA and protein was evident in the basolateral amygdala (BLA) of Lmo3Z mice. To examine whether Lmo3 in the amygdala is important for anxiety-like behavior, we locally reduced Lmo3 expression in the BLA of wild type mice using a lentiviral vector expressing a short hairpin RNA targeting the Lmo3 transcript. Mice with Lmo3 knockdown in the BLA exhibited decreased anxiety-like behavior relative to control mice. These results suggest that Lmo3 promotes anxiety-like behavior specifically in the BLA, possibly by altering Crhr1 expression. This study is the first to support a role for Lmo3 in anxiety-like behavior.
Keywords: LMO3, anxiety, CRF, CRH, CRF1 receptor, basolateral amygdala
1. Introduction
Anxiety disorders carry a heavy cost, both at the personal and at the societal level, and this cost is made greater due to the high prevalence of these disorders (Konnopka et al., 2009). Although anxiety disorders are highly heritable and several genes have been associated with their occurrence (Lacerda-Pinheiro et al., 2014), few genes have been identified that have proven to aid in treatment outcomes (Serretti et al., 2009). There is, therefore, a great need to identify genes that confer either susceptibility to or resilience from anxiety. Research into the neurobiology of anxiety has provided insight into its mechanisms. Altering signaling in the amygdala, an area of the brain that is primarily known for its regulation of fear conditioning and processing of emotional salience, can produce changes in anxiety-like behavior (Calhoon and Tye, 2015; Sanders and Shekhar, 1995). The neuropeptide corticotropin releasing factor (CRF, also known as corticotropin releasing hormone, encoded by the Crh gene) has emerged as a key regulator of amygdalar signaling (Gray et al., 2015; Silberman and Winder, 2013) and anxiety (Shekhar et al., 2005). CRF binds to two receptors, and its actions at the CRF receptor 1 (CRHR1, encoded by the Crhr1 gene) appear to be particularly critical to its effects on anxiety. Mice lacking the Crhr1 gene show a decreased anxiety-like phenotype (Contarino et al., 1999; Muller et al., 2003; Smith et al., 1998; Timpl et al., 1998), and CRHR1 antagonists administered systemically (Zorrilla et al., 2002) or directly into the amygdala (Cipriano et al., 2016; Sotnikov et al., 2014) reduce anxiety-like behavior. Although the role of CRHR1 in regulating anxiety is well established, the mechanisms regulating CRHR1 expression are still largely unknown. Therefore, identifying genes that may alter amygdalar signaling and/or CRHR1 expression could provide potential new targets for the development of effective pharmacotherapies for anxiety.
LMO3 is one of four LMO proteins that are unique transcriptional regulators involved in cell differentiation and behavior (Zheng and Zhao, 2007). LMO proteins do not bind to DNA but instead interact with transcription factors to repress or activate transcription, a regulatory mechanism for controlling the cell-type or brain-region specificity of transcription factor activity (Bifsha et al., 2016). LMO3 is important for the differentiation of interneurons into specific subtypes (Au et al., 2013), but it is not necessary for survival. Lmo3 knockout (Lmo3Z) mice are viable and fertile, with no obvious health problems or gross morphological brain defects (Tse et al., 2004). Although LMO3 is ubiquitously expressed throughout the brain, it shows particularly dense expression within subcortical regions, including the amygdala (Hinks et al., 1997).
Several LMO proteins have also been associated with drug- and alcohol-related phenotypes, in Drosophila (Lasek et al., 2011), mice (Lasek et al., 2010), and humans (Kapoor et al., 2013). LMO3, specifically, has been shown to regulate alcohol consumption and sensitivity to the sedative effects of alcohol (Lasek et al., 2011; Savarese et al., 2014), with loss of Lmo3 in mice increasing binge-like alcohol consumption (Savarese et al., 2014). Although the role of LMO3 in regulating other behavioral phenotypes has not yet been studied, another related LMO protein, LMO4, regulates anxiety (Qin et al., 2015) and fear learning (Maiya et al., 2012) via its actions in the BLA. Given the high comorbidity of alcohol abuse and anxiety (Kushner et al., 1990; Merikangas et al., 1998; Regier et al., 1990) and high expression of LMO3 in the amygdala (Hinks et al., 1997), we hypothesized that Lmo3 may be a novel regulator of anxiety.
Here, we examined whether LMO3 regulates anxiety-like behavior utilizing the Lmo3Z mouse in two well-validated measures of anxiety-like behavior: the elevated plus maze (EPM) and the novelty-induced hypophagia (NIH) tests. Additionally, we examined whether Lmo3 may regulate expression of genes that are known to be critical for anxiety, namely Crh and Crhr1. Finally, to test if Lmo3 expression in the amygdala is important for anxiety-like behavior, we knocked-down Lmo3 in the amygdala of wild-type mice using a lentiviral vector expressing a short hairpin RNA (shRNA) targeting Lmo3, and tested mice on the EPM. Our results indicate a role for Lmo3 in promoting anxiety-like behavior and Crhr1 gene expression, specifically via its actions in the BLA.
2. Material and methods
2.1. Subjects
Lmo3Z mice containing an IRES-LacZ insertion in exon 2 of Lmo3 have been described previously (Savarese et al., 2014; Tse et al., 2004). Mice were previously backcrossed 2 generations onto the C57BL/6J background for behavioral testing (Savarese et al., 2014). Adult (~10–16 weeks old) male and female homozygous Lmo3Z and wild-type littermates were used for behavioral testing and gene expression experiments. Male and female C57BL/6J mice (8 weeks old at the time of purchase and 8–10 weeks old at the time of surgery) were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice were group housed (except during the NIH task) with same-sex cage mates in a temperature- and humidity-controlled environment under a 14-hour light/dark cycle (lights on at 6 am and off at 8 pm). All behavioral testing took place in the morning, approximately four hours into the light phase. Mice had access to food and water ad libitum for the duration of the study and were maintained and cared for in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All procedures involving animals were approved by the University of Illinois at Chicago Animal Care Committee.
2.2. EPM
The EPM is widely used as a measure of anxiety-like behavior in rodents. The maze itself consisted of four arms (two open and two enclosed by 12 cm high walls) that were 45 cm long and 10 cm wide, elevated approximately 50 cm above the ground. At the start of a trial, the mouse was placed into the center of the maze with its nose directed at one of the closed arms and allowed to freely explore for 5 minutes. The total distance traveled, as well as the amount of time spent in each arm and the number of entries into each arm were digitally tracked and recorded. Additional parameters determined in data analysis included percent entries into each arm relative to total entries.
2.3. NIH
NIH is a well-documented phenomenon occurring in rodents and it can be observed without the use of food restriction (Dulawa, 2007). Importantly, when utilized as a behavioral measure in the laboratory, NIH exhibits strong predictive validity as a measure of anxiety. Most of the traditional anxiety measures for mice depend on locomotion (i.e., the open field and light-dark box), but NIH uses within-subjects comparisons for each outcome measure, allowing for any differences in activity to be controlled (i.e., if there is reduced locomotion that is independent of anxiety-like behavior, it will be observed in both the familiar context (home cage) and the novel context and will not impact primary outcome measures). Mice were singly housed for 3 days prior to and for the duration of testing in this task. Water bottles were removed from cages and mice received sipper tubes of diluted sweetened condensed milk (Eagle Brand, diluted in water 1:3) for 30 minutes across 3 training days. The amount of milk consumed in each session was recorded to ensure that groups did not differ in consumption levels by the end of training. On the fourth day (home-cage testing), testing was nearly identical to training days, except that the latency to the first lick of milk was measured, and consumption was measured every 5 minutes throughout the 30-minute period. On the fifth day, mice were placed in a novel context (a new, clean cage with bedding removed) for testing. The cage was placed on white paper and a light was shone overhead to enhance the anxiogenic environment. Once again, mice received the sipper tubes containing the sweetened milk solution for 30 minutes and latency and consumption were recorded as in the home-cage test environment.
2.4. Quantitative real-time polymerase chain reaction (qPCR)
For gene expression experiments, Lmo3Z and wild-type mice were euthanized by CO2 inhalation and rapidly decapitated. Brains were removed, rinsed in cold PBS, and sectioned on ice into 1 mm-thick coronal sections using an adult mouse brain matrix (Zivic Instruments, Pittsburgh, PA, USA), from which individual brain areas were punched from the tissue sections using disposable glass Pasteur pipettes. Amygdala punches were obtained in 1 mm-thick sections at approximately −0.8 to −1.8 mm posterior to bregma. The BLA was punched between the branches of the external capsule and the CeA was punched immediately medial to the BLA. Tissue was immediately frozen on dry ice in 1.5 mL centrifuge tubes and stored at −80°C. The procedure for tissue collection in mice that received the lentiviral injections was slightly modified. After euthanasia, whole brains were removed, rinsed in cold PBS, and immediately placed into a 12-well plate on dry ice and stored at −80°C. Brains were sectioned to 300 μm thickness and mounted onto glass slides resting on dry ice. Fluorescent tissue was collected in the dark, on dry ice, using a Dual Fluorescent Protein Flashlight (Nightsea, Bedford, MA, USA) and 1 mm-diameter biopsy punches (Integra LifeSciences, Plainsboro, NJ, USA) at approximately −1.6 mm posterior to bregma. Tissue punches were immediately frozen in 1.5 ml centrifuge tubes and stored at −80°C until processed. RNA was isolated using the GeneJET RNA Purification kit (Thermo Fisher Scientific) and cDNA was synthesized from equal amounts of input RNA using the Maxima First Strand cDNA Synthesis kit for RT-qPCR (Thermo Fisher Scientific). qPCR was performed using either Maxima SYBR Green or Maxima Probe qPCR Master Mix (Thermo Fisher Scientific), Crh, Crhr1, Lmo3, and Gusb primers (Crh forward, 5′-CAACCTCAGCCGGTTCTGAT; Crh reverse, 5′-CAGCGGGACTTCTGTTGAGA; Crhr1 forward, 5′-TCCCCATCATTGTGGCTTGG; Crhr1 reverse, 5′-ATCATGGGGCCCTGGTAGAT; Lmo3 forward, 5′-TGCGACCGCATAAAGGTGAG; Lmo3 reverse, 5′-GCACAACCTTTTGGCTTGGT; Gusb forward, 5′-CGGGACTTTATTGGCTGGGT; Gusb reverse, 5′-CCATTCACCCACACAACTGC), and 20X pre-made FAM-labeled mouse Actb probe/primer mix (Life Technologies, Carlsbad, CA, USA). Relative expression of Crh, Crhr1, and Lmo3 was calculated using the 2−ΔΔCq method using Gusb or Actb expression as a reference as indicated in each figure. Expression was normalized to the average ΔCq in the BLA of control mice (Livak and Schmittgen, 2001).
2.5. Western blotting
Tissue was collected in the same manner as described for qPCR analysis (above). Tissue was homogenized in 100 μl of RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 ug/mL leupeptin) supplemented with protease inhibitors (Halt Protease Inhibitor Cocktail, Thermo Scientific). Protein concentrations were determined using the BCA Protein Assay Kit (Fisher Scientific, Pittsburgh, PA, USA). Equal amounts of protein (30 μg) were subjected to polyacrylamide gel electrophoresis (Novex WedgeWell 10% Tris-Glycine Gel, Invitrogen, Thermo Fisher Scientific) and transferred to PVDF membranes. Membranes were blocked with 5% milk (for β-Actin) or 5% BSA (for CRHR1) in TBST (25 mM Tris-HCl, 137 mM NaCl and 0.1% Tween 20) and incubated with primary antibodies overnight at 4°C (goat polyclonal anti-CRF1 [CRHR1], 1:500, Abcam, ab59023; mouse monoclonal anti-β-Actin, Sigma Aldrich A5441, 1:10,000). The membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-mouse IgG, 1:3000 for CRHR1 or rabbit anti-goat, 1:1000 for β-actin; Bio-Rad, Hercules, CA) at room temperature for 90 minutes and developed with enhanced chemiluminescence (ECL) detection reagents (Pierce ECL Western Blotting Substrate, Thermo Scientific). Band intensities were quantified using NIH Image J software and protein levels were normalized to β-Actin protein for each sample.
2.6. Lentiviral vectors
Lentivirus expressing short hairpin RNAs (shRNAs) targeting Lmo3 (shLmo3) or a nonspecific sequence not known to target any gene in the mouse genome (shScr) were described previously (Lasek et al., 2011). The shRNAs are expressed from the U6 promoter and enhanced green fluorescent protein (GFP) is expressed from a CMV promoter.
2.7. Stereotaxic surgery
Male and female mice (8–10 weeks old) were anesthetized with ketamine (100 mg/kg, i.p.) and xylazine (8 mg/kg, i.p.) and placed in a digital stereotaxic alignment apparatus (Model 1900, David Kopf Instruments, Tujunga, CA, USA). After identification of bregma and leveling of the skull, 0.28 mm diameter holes were drilled bilaterally (A/P: −1.6, M/L: ±3.1) for microinjections of virus. Mice were randomized to receive either shLmo3 or shScr. Cannulas (33 gauge) were inserted bilaterally into the basolateral amygdala (D/V: −4.8) and 1 μl of virus was infused per side at a rate of 0.2 μl per minute. Viral titers were ~3 × 107 pg/mL gag antigen as measured by ELISA. Mice received a single injection of meloxicam (2 mg/kg, subcutaneously) post-surgery for analgesia. Behavioral testing began 3 weeks after infection. A separate group of mice did not undergo behavioral testing, but were instead used to verify in vivo knockdown of Lmo3 using qPCR. These mice were allowed to recover for 3 weeks after surgery before they were euthanized and the BLA dissected and analyzed as described above.
2.8 Immunohistochemistry
In order to verify infection in the BLA, mice were transcardially perfused with PBS followed by 4% paraformaldehyde. Brains were removed and post-fixed overnight at 4°C in 4% paraformaldehyde, and were then transferred to 30% sucrose in PBS for an additional 24 hours. Brains were mounted with optimal cutting temperature (OCT) media for sectioning on the cryostat and were cut to 50 μm free-floating sections in PBS. The sections were treated with 3% H2O2 for 10 minutes followed by 50% ethanol twice, each time for 10 minutes. 10% normal donkey serum in 0.25% Triton was used for blocking (30 minutes) and sections were then incubated with mouse anti-GFP monoclonal antibody (Life Technologies, A11120) diluted 1:1000 in 2% normal donkey serum in PBS overnight at 4°C. Sections were then incubated with biotin-conjugated horse anti-mouse secondary antibody (1:200, Vector Laboratories, Burlingame, CA, USA, BA-2000) for 1 hour at room temperature followed by ABC-Peroxidase solution (Vectastain ABC kit, Vector Laboratories, Burlingame, CA, USA, PK-6100) for an hour. Diaminobenzidine (DAB) peroxidase substrate (Vector Laboratories, Burlingame, CA, USA) was then applied for ~1 minute for brown color detection of the GFP immunostaining. Sections were mounted on gelatin-coated slides and allowed to dry. Counterstaining with cresyl violet was performed and slides were coverslipped with Permount. Mice were only included in the data analysis if the infection was bilateral and clearly localized only to the BLA.
2.8. Statistical analysis
Unpaired student’s t-tests were employed to evaluate EPM and gene expression (qPCR and Western Blot) data between genotypes. A Welch’s correction was applied if the variance differed between groups. Two-way repeated measures analysis of variance (RM ANOVA) was utilized for evaluation of NIH data followed by Sidak’s post hoc comparisons as appropriate. Sex was first evaluated as an independent variable for all analyses. In all experiments, no main effects of sex or interactions were observed, so the data were collapsed across sex and then tested for genotype or group differences. Graphs show the male and female data combined. All statistical analyses were performed using GraphPad Prism software version 6.05 (GraphPad, La Jolla, CA, USA).
3. Results
3.1. Lmo3Z mice show decreased anxiety in the EPM
To determine if Lmo3 regulates anxiety-like behavior, we compared Lmo3Z mice to their wild-type littermates in the EPM. Lmo3Z mice spent more time in the open arms (Fig. 1A) [t(45) = 2.23, p = 0.031] and had a greater percentage of entries into the open arms relative to total entries compared with control mice (Fig. 1B) [t(45) = 4.01, p = 0.0002]. Because mice tend to avoid the open arms due to an innate fear of open spaces and heights, an increase in open arm time is interpreted as a reduction in anxiety-like behavior. However, open arm time is dependent on locomotor activity, and Lmo3Z mice demonstrated reduced activity relative to control mice as measured by total arm entries and distance traveled (Fig. 1C, D) [entries, t(45) = 10.99, p < 0.0001; distance, t(45) = 14.59, p < 0.0001]. Although the increase in open arm time and entries by Lmo3Z mice is consistent with reduced anxiety, these results are confounded by the decreased overall activity of the Lmo3Z mice relative to controls.
Fig. 1. Lmo3Z mice show decreased anxiety in the EPM.
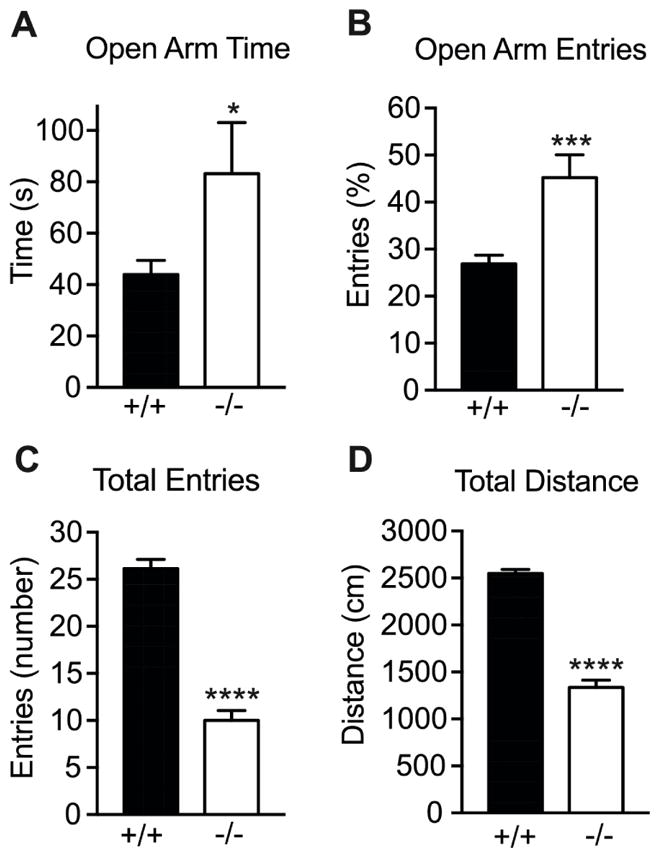
Homozygous Lmo3Z mice (−/−, white bars, n = 19) and wild-type littermates (+/+, black bars, n = 28) were tested in the elevated plus maze. A) Open arm time. B) Percentage of open arm entries. C) Total entries into open and closed arms. D) Total distance traveled. *p < 0.05, ***p < 0.001, and ****p < 0.0001 as measured by student’s t-test.
3.2. Lmo3Z mice do not exhibit NIH
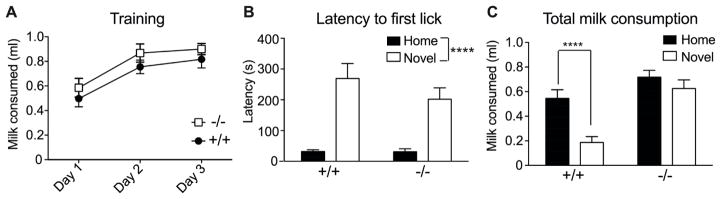
We next sought an anxiety measure that was independent of locomotor activity in order to confirm whether Lmo3Z mice exhibit decreased anxiety-like behavior, so mice were tested for NIH. Consumption of the sweetened milk solution did not differ between genotypes across the training days [F(1,30) = 1.31, p = 0.26), but there was a significant effect of time, with both Lmo3Z and wild-type mice consuming more milk solution across the training period (Fig. 2A) [time: F(2,60) = 40.54, p < 0.0001]. Genotypes also did not differ in the latency to the first lick [F(1,30) = 1.02, p = 0.32]. While both Lmo3Z and wild-type mice exhibited an increased latency to the first lick in the novel context relative to the familiar context (home cage), they did so to the same extent (Fig. 2B) [context: F(1,30) = 41.75, p < 0.0001]. However, there was a significant main effect of genotype and a context by genotype interaction for total milk consumption (Fig. 2C) [context: F(1,30) = 51.96, p < 0.0001; genotype: F(1,30) = 13.74, p = 0.0008; interaction: F(1,30) = 17.99, p = 0.0002]. Post-hoc multiple comparisons tests indicated that while the wild-type mice showed the expected reduction of feeding in the novel context relative to the home cage (p < 0.0001), the Lmo3Z mice did not (p = 0.111). The failure of the Lmo3Z mice to reduce their milk consumption in the novel environment suggests that they are less sensitive to the anxiety-producing effects of this environment. These data, together with the EPM findings, indicate that Lmo3Z mice have reduced levels of anxiety relative to wild-type littermates.
Fig. 2. Lmo3Z mice do not exhibit novelty-induced hypophagia.
Homozygous Lmo3Z mice (−/−, n = 14) and wild-type littermates (+/+, n = 18) were tested for novelty-induced hypophagia. A) Sweetened condensed milk consumption on the training days. B) Latency to first lick in the home cage (black bars) vs. the novel context (white bars). C) Total milk consumption in the home cage (black bars) vs. the novel context (white bars). Note that the Lmo3Z mice consumed similar amounts of milk in the novel context and home cage. ****p < 0.0001 by two-way RM ANOVA (B) or Sidak’s post hoc multiple comparisons test (C).
3.3. Reduced Crhr1 mRNA and protein expression in the BLA of Lmo3Z mice
Lmo3 expression is particularly dense in subcortical areas of the brain, including the BLA (Hinks et al., 1997; Savarese et al., 2014). Because the BLA plays such a prominent role in regulating anxiety, we next tested whether LMO3 may be regulating the expression of genes in the BLA that underlie anxiety-like behavior – i.e., Crh and Crhr1. To do this, the BLA and CeA were dissected from the brains of Lmo3Z and wild-type mice and expression of Crh and Crhr1 were measured by qPCR. Crhr1 levels were significantly reduced by ~30% in Lmo3Z relative to wild-type mice in both the BLA (Fig. 3A) [t(44) = 2.88, p = 0.0061] and the CeA (Fig. 3C) [t(36) = 2.31, p = 0.027]. Crhr1 mRNA levels were slightly higher in BLA than in CeA in wild type mice, consistent with published findings (Van Pett et al., 2000). Crh expression in the BLA (Fig. 3B) and CeA (Fig. 3D) was not altered in Lmo3Z compared with control mice [BLA: t(44) = 1.165, p = 0.25; CeA: t(36) = 0.67, p = 0.51], nor did we observe a difference in mRNA levels between BLA and CeA. Mean Cq values for the reference gene, Actb did not differ between genotypes in either the BLA [wild-type, 19.85 ± 0.09; Lmo3Z, 19.74 ± 0.15; t(44) = 0.65, p = 0.52] or the CeA [wild-type, 19.65 ± 0.10; Lmo3Z, 19.50 ± 0.11; t(36) = 0.97, p = 0.34].
Fig. 3. Reduced Crhr1 mRNA expression in the BLA and CeA of Lmo3Z mice.
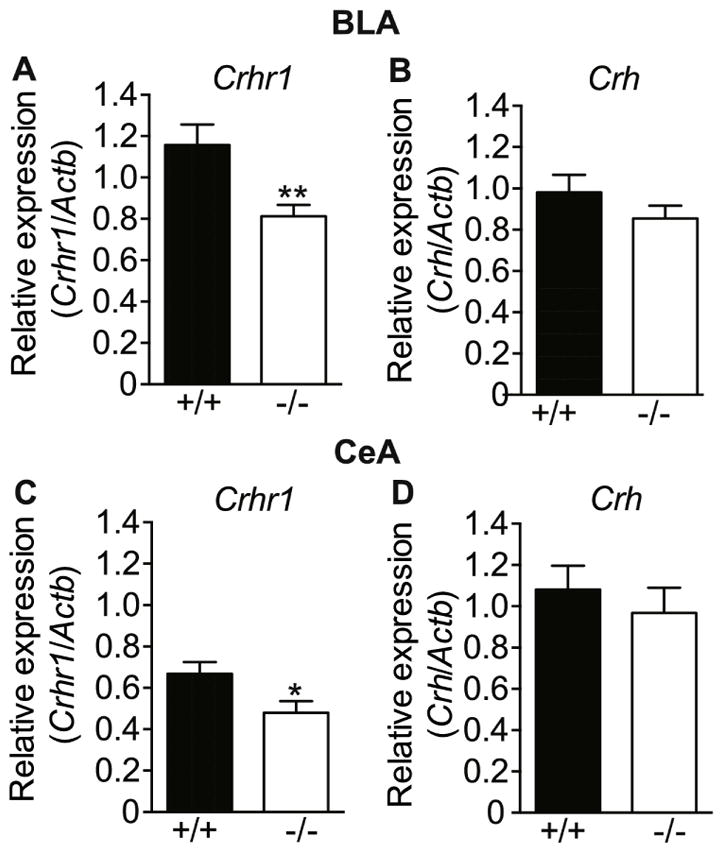
Basolateral (BLA) and central (CeA) amygdala were dissected from the brains of Lmo3Z (−/−, white bars) and wild-type (+/+, black bars) mice for qPCR analysis of mRNA levels. Crhr1 expression in the A) BLA (n = 21 Lmo3Z and 25 wild-type) and C) CeA (n = 17 Lmo3Z and 21 wild-type), Crh expression in the B) BLA (n = 22 Lmo3Z and 24 wild-type) and D) CeA (n = 17 Lmo3Z and 21 wild-type). *p<0.05, **p < 0.01 by student’s t-test.
To determine if the reduction in Crhr1 also resulted in a decrease in the encoded protein, the BLA and CeA of Lmo3Z and wild-type mice were analyzed for CRHR1 by Western blotting. Expression of CRHR1 was also significantly reduced by 45% in the BLA of Lmo3Z mice compared with wild-type mice (Fig. 4A) [t(19) = 2.20, p = 0.04], whereas there was no change in CRHR1 protein in the CeA of Lmo3Z mice (Fig. 4B) [t(23) = 0.83, p = 0.42]. β-actin band intensity did not differ between genotypes in the BLA [wild-type, 5999 ± 779; Lmo3Z, 6621 ± 409; t(19) = 0.64, p = 0.53] or in the CeA [wild-type, 9388 ± 1016; Lmo3Z, 9638 ± 848; t(23) = 0.19, p = 0.85]. These results suggest that Lmo3 regulates the expression of the Crhr1 gene, with loss of Lmo3 resulting in reductions of CRHR1 mRNA and protein in the BLA.
Fig. 4. Reduced CRHR1 protein in the BLA of Lmo3Z mice.
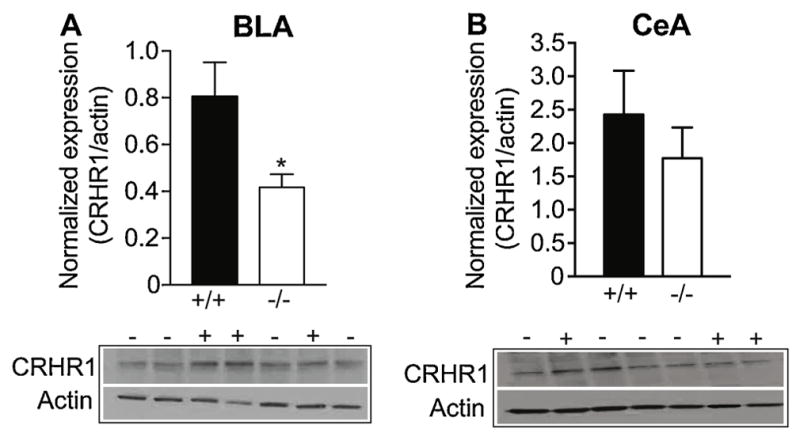
Basolateral (BLA) and central (CeA) amygdala were dissected from the brains of Lmo3Z (−/−, white bars) and wild-type (+/+, black bars) mice for Western Blot analysis. Graphs show the quantification of CRHR1 expression in the A) BLA (n = 9 Lmo3Z and 12 wild-type ) and B) CeA (n = 13 Lmo3Z and 12 wild-type) relative to actin. Representative CRHR1 and actin western blots are shown below each graph. *p < 0.05 by student’s t-test.
3.4. Knockdown of Lmo3 in the BLA reduces anxiety-like behavior
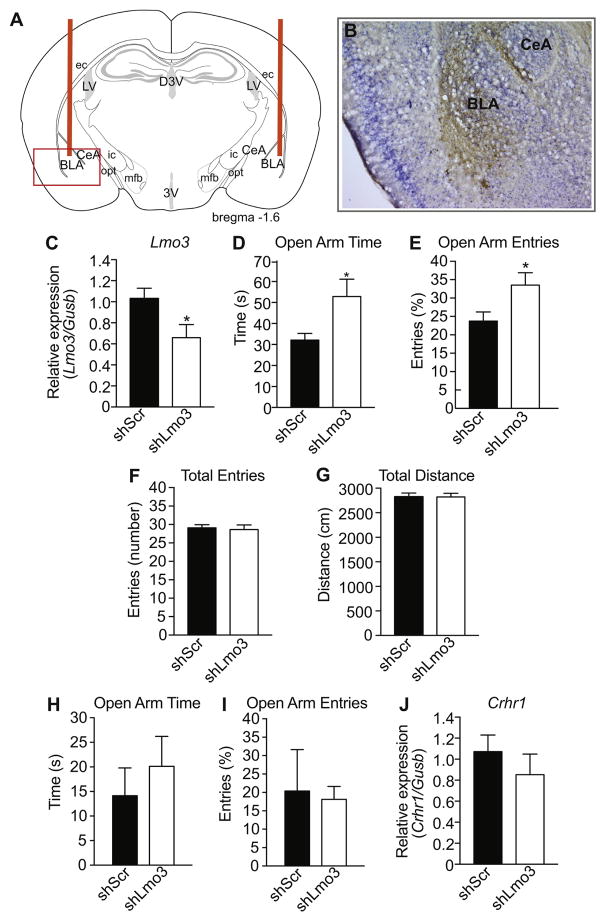
To determine whether the anxiolytic phenotype observed in Lmo3Z mice was due to Lmo3 expression in the BLA, we utilized viral-mediated RNA interference to knockdown expression of Lmo3 in the mouse BLA. Mice were stereotaxically injected in the BLA with a lentivirus expressing a non-targeting control shRNA (shScr) or an shRNA targeting Lmo3 (shLmo3) (Lasek et al., 2011). Viral infection in the BLA of all mice was verified by immunohistochemistry using an antibody to GFP after the completion of behavioral testing, an example of which is shown in Fig. 5A–B. We also measured Lmo3 expression in the BLA by qPCR in a separate group of mice to confirm the effectiveness of the vector. Lmo3 was reduced by 35% in the BLA of mice expressing shLmo3 compared with shScr (Fig. 5C) [t(15) = 2.32, p = 0.035). Cq values of the reference gene, Gusb, did not differ between groups [wild-type, 30.36 ± 0.24; Lmo3Z, 29.86 ± 0.46; t(15) = 0.93, p = 0.37]. Mice expressing shLmo3 spent more time in the open arms (Fig 5D) [t(21) = 2.1, p = 0.048] and had an increased percentage of open arm entries (Fig. 5E) [t(21) = 2.20, p = 0.039]. Knockdown of Lmo3 in the BLA had no effect on the total arm entries [t(21) = 0.30, p = 0.77] or the total distance traveled [t(21) = 0.051, p = 0.96] (Fig. 5F–G), demonstrating that adult-specific knockdown of Lmo3 in the BLA does not affect overall locomotor activity, in contrast to what we observed with Lmo3Z mice. Additionally, the anxiolytic phenotype produced by Lmo3 knockdown appears to be specific to the BLA. In mice that had off-target infection of lentivirus in the nearby CeA, Lmo3 knockdown had no effect on EPM behavior (Fig. 5H–I) [open arm time: t(4) = 0.72, p = 0.51; percent open arm entries: t(4) = 0.19, p = 0.86]. Together, these data indicate that Lmo3 expression in the BLA of adult mice is important for promoting anxiety-like behavior.
Fig. 5. Knockdown of Lmo3 in the basolateral amygdala (BLA) reduces anxiety-like behavior in the elevated plus maze (EPM).
Mice were infected in the BLA with a lentivirus expressing a control shRNA (shScr, n = 10) or an shRNA targeting Lmo3 (shLmo3, n = 13) and tested in the EPM three weeks after injection. A) Schematic of coronal brain section 1.6 mm posterior to bregma showing cannula insertion for injection (orange bars). The red box highlights the area of the photograph shown in B). B) Representative image of infection as exhibited by GFP immunohistochemistry (brown staining). Blue color is cresyl violet counterstain. C) Lmo3 expression as measured by qPCR in a separate group of mice than those that were tested in the EPM (n = 9 shLmo3 and 8 shScr). D) Open arm time. E) Percent entries into the open arm. F) Total number of arm entries. G) Total distance traveled. H) Open arm time of mice that were infected with lentivirus in the CeA. I) Percent entries into the open arm of mice that were infected with lentivirus in the CeA. J) Crhr1 expression in the BLA of the same mice used in C) to verify Lmo3 knockdown. *p < 0.05 by student’s t-test. Abbreviations: BLA, basolateral amygdala; CeA, central amygdala; LV, lateral ventricle; D3V, dorsal part of 3rd ventricle; 3V, 3rd ventricle; ec, external capsule; ic, internal capsule; mfb, medial forebrain bundle, opt, optic tract.
To test if Lmo3 knockdown in the BLA altered Crhr1 expression, we also measured Crhr1 expression by qPCR in the same animals that were used to verify in vivo knockdown of Lmo3 (Fig. 5C). No significant difference in Crhr1 expression was observed between mice expressing shLmo3 compared with those expressing shScr in the BLA [t(13) = 0.86, p = 0.41] (Fig. 5J).
4. Discussion
In this study we describe a novel role for Lmo3 in the control of anxiety-like behavior, specifically through its actions in the BLA. We demonstrate that Lmo3 knockout mice display decreased anxiety-like behavior that can be replicated by local knockdown of Lmo3 in the BLA of adult wild-type mice. The CeA is also important for anxiety-like behavior (Gilpin et al., 2015), but knockdown of Lmo3 in the CeA did not affect anxiety-like behavior, demonstrating anatomical specificity in the action of LMO3 in the amygdala with regard to this behavioral measure. Together, these results suggest that LMO3 plays an active role in the BLA of adult animals to promote anxiety-like behavior.
LMO3 is important for the differentiation of cortical GABA interneurons into the parvalbumin-expressing subtype during development (Au et al., 2013); however, our data indicates that the role of Lmo3 in anxiety-like behavior is probably not due to a developmental effect on neuronal differentiation because knockdown of Lmo3 in adult animals recapitulates the phenotype observed in the knockout animals.
Here, we attempted to identify a potential downstream target of LMO3 that regulates anxiety. We detected a significant reduction in Crhr1 mRNA and protein expression in the BLA of Lmo3Z mice relative to wild-type mice, suggesting that Lmo3 may promote transcription of the Crhr1 gene in the BLA. It is likely that the decrease in Crhr1 in the BLA of Lmo3Z mice contributes to decreased anxiety-like behavior, because Crhr1 knockout mice exhibit reduced anxiety-like behavior (Muller et al., 2003; Refojo et al., 2011; Smith et al., 1998; Timpl et al., 1998) and injection of selective CRHR1 antagonists, CP376395 or α-helical CRF 9–41, into the BLA of mice decreases anxiety-like behavior (Cipriano et al., 2016; Sotnikov et al., 2014). Moreover, lentiviral shRNA-mediated knockdown of Crhr1 in the mouse BLA decreases anxiety (Sztainberg et al., 2010). Interestingly, we found that Crhr1 transcript was also decreased in the CeA of Lmo3Z mice, but this did not translate to a reduction in protein levels. More detailed mechanistic studies are warranted to determine how LMO3 regulates transcription of the Crhr1 gene in the amygdala and whether increasing expression of Crhr1 in the BLA of Lmo3Z mice would increase anxiety-like behavior in these mice.
Another mechanism for the regulation of CRHR1 expression is by CRH, since CRH can downregulate the expression of CRHR1 in the hippocampus and cortex (Brunson et al., 2002). It is possible that loss of LMO3 results in increased CRH and thus decreased CRHR1 in the amygdala. However, no differences were found in Crh mRNA levels in the BLA or CeA of Lmo3Z mice compared with wild type mice. We can’t rule out the possibility that there are alterations in CRH protein levels in the amygdala, since we only examined transcript, but even if CRH protein levels in the amygdala were elevated in Lmo3Z mice, CRH apparently does not cause a downregulation of CRHR1 in this brain region (Brunson et al., 2002).
Although knockdown of Lmo3 in the BLA of adult mice using a virus-delivered shRNA targeting Lmo3 resulted in decreased anxiety-like behavior, we did not observe a significant decrease in Crhr1 mRNA expression in the BLA of mice expressing shLmo3. There are a few potential explanations for this result. First, it is entirely possible that for technical reasons we could not reliably detect changes in Crhr1 expression in mice infected with lentivirus expressing shLmo3 in the BLA. Although we attempted to carefully dissect only fluorescent (i.e. infected) BLA tissue, there were likely cells in the dissected tissue that were not infected, which would minimize the extent of knockdown. In fact, we only observed a 35% knockdown of Lmo3 in the BLA, whereas Lmo3 is completely absent in the Lmo3Z mice (in which Crhr1 BLA expression is decreased by 30%). Injection of a higher volume or titer of lentivirus could potentially increase the amount of infection and Lmo3 knockdown, but increased injection volume is expected to damage the amygdala. The lentiviral titer achieved is also limited by the volume in which the virus can be resuspended after concentrating. Thus, it is possible that Crhr1 expression was significantly altered in cells expressing shLmo3 but we could not detect this decrease using the available methods. Alternatively, it may be that the Lmo3 knockdown altered CRHR1 protein levels or functionality without changing Crhr1 transcript levels. Finally, it is still a possibility that LMO3 might be acting independently of Crhr1 to promote anxiety-like behavior.
To date, little is known regarding potential target genes and signaling pathways regulated by LMO3, especially in the nervous system. LMO3 can promote or repress gene transcription, depending on the cellular context (Isogai et al., 2011; Larsen et al., 2010; Olsavszky et al., 2017; Susa et al., 2009). LMO3 has been shown to cooperate with pituitary homeobox 3 (PITX3) to regulate its own expression (Bifsha et al., 2016), as well as to interact with the tumor suppressor p53 to inhibit its transcriptional targets (Larsen et al., 2010). Interestingly, LMO3 has also been shown to cooperate with the transcription factor PPARγ (Lindroos et al., 2013). Neuron-specific deletion of PPARγ increases anxiety-like behavior (Domi et al., 2016). This raises the possibility that LMO3 could potentially increase anxiety by inhibiting PPARγ. Based on these findings, one would predict that Lmo3 knockout mice have elevated PPARγ activity. Future experiments are needed to test this hypothesis.
Anxiety and alcohol use often co-occur in humans, and the direction of this comorbidity is typically positive, such that either high anxiety promotes alcohol drinking or withdrawal from alcohol use leads to increased anxiety. We previously found that Lmo3Z mice engage in elevated binge-like drinking compared to controls (Savarese et al., 2014), yet here we show that these mice exhibit a reduced anxiety phenotype. This dissociation between high anxiety and elevated drinking behavior is not unique (Hwang et al., 2004; Tuominen et al., 1990)(Acewicz et al., 2014). The high-drinking-in-the-dark (HDID) mouse lines exhibit either no difference in anxiety-like behavior or a slight reduction in basal anxiety levels (Barkley-Levenson and Crabbe, 2015), and the high-alcohol-preferring (HAP) mouse lines either exhibit reduced anxiety or no difference in anxiety, depending on the test (Can et al., 2012). One explanation for the reduced anxiety but high alcohol consumption phenomenon could be an increase in risk-taking or exploratory behavior. It is possible that the Lmo3Z mice engage in elevated binge drinking because of increased novelty seeking, risk-taking, or increased sensitivity to alcohol reward. Additional behaviors in Lmo3Z mice related to these endophenotypes should be examined to further define the role of LMO3 in addiction and mood disorders.
Another gene in the LMO family, Lmo4, also regulates anxiety-like behavior. Mice with Lmo4 deleted in glutamatergic neurons exhibit increased anxiety-like behavior (Qin et al., 2015). This is particularly intriguing in that it suggests potential opposing roles of LMO3 and LMO4 in the regulation of anxiety, with LMO3 promoting, and LMO4 inhibiting anxiety. It is currently not known which cell types in the BLA express LMO3. However, conditional knockout of Crhr1 in mouse glutamatergic forebrain neurons decreases anxiety (Refojo et al., 2011), consistent with our results demonstrating that Lmo3Z mice exhibit decreased anxiety-like behavior and Crhr1 expression. We speculate that LMO3 acts in glutamatergic neurons of the BLA to regulate transcription of Crhr1 and thus anxiety-like behavior based on these findings. Future studies should delineate the cell types in which LMO3 acts to promote anxiety and Crhr1 expression.
5. Conclusions
This is the first study to explore the role of the Lmo3 gene in regulating anxiety, and particularly how this regulation of behavior could be driven by the actions of Lmo3 in a distinct brain region and via regulation of a specific gene. Taken together, the current data highlight a novel role for Lmo3 in promoting anxiety-like behavior via its actions in the BLA. Lmo3Z mice show a reduction of Crhr1 expression in the BLA that may be indicative of altered signaling in this brain region. These results suggest that Lmo3 may be a novel contributor to the etiology of anxiety disorders, and better understanding its role in the regulation of amygdala signaling and stress-related networks could provide new discoveries for effective therapeutics.
Highlights.
LIM domain only 3 (Lmo3) knockout mice display reduced anxiety-like behavior.
Lmo3 knockout mice have decreased Crhr1 expression in the basolateral amgydala (BLA).
Knockdown of Lmo3 in the BLA of wild-type mice using viral-mediated RNA interference reduces anxiety.
High expression of Lmo3 may be a novel genetic risk factor for anxiety.
Acknowledgments
Funding: This work was supported by grants from the National Institutes of Health (P50 AA022538, U01 AA020912, and R01 DA033429).
The authors declare no conflicts of interest.
Footnotes
Declarations of Interest: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acewicz A, Mierzejewski P, Jastrzebska A, Korkosz I, Karas K, Sienkiewicz-Jarosz H, Samochowiec J, Kolaczkowski M, Bienkowski P. Anxiety- and depressive-like traits in Warsaw alcohol high-preferring (WHP) and Warsaw alcohol low-preferring (WLP) rats. Pharmacol Biochem Behav. 2014;122:261–265. doi: 10.1016/j.pbb.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Au E, Ahmed T, Karayannis T, Biswas S, Gan L, Fishell G. A modular gain-of-function approach to generate cortical interneuron subtypes from ES cells. Neuron. 2013;80:1145–1158. doi: 10.1016/j.neuron.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley-Levenson AM, Crabbe JC. Genotypic and sex differences in anxiety-like behavior and alcohol-induced anxiolysis in High Drinking in the Dark selected mice. Alcohol. 2015;49:29–36. doi: 10.1016/j.alcohol.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bifsha P, Balsalobre A, Drouin J. Specificity of Pitx3-Dependent Gene Regulatory Networks in Subsets of Midbrain Dopamine Neurons. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0040-y. [DOI] [PubMed] [Google Scholar]
- Brunson KL, Grigoriadis DE, Lorang MT, Baram TZ. Corticotropin-releasing hormone (CRH) downregulates the function of its receptor (CRF1) and induces CRF1 expression in hippocampal and cortical regions of the immature rat brain. Exp Neurol. 2002;176:75–86. doi: 10.1006/exnr.2002.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoon GG, Tye KM. Resolving the neural circuits of anxiety. Nat Neurosci. 2015;18:1394–1404. doi: 10.1038/nn.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can A, Grahame NJ, Gould TD. Affect-related behaviors in mice selectively bred for high and low voluntary alcohol consumption. Behav Genet. 2012;42:313–322. doi: 10.1007/s10519-011-9505-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriano AC, Gomes KS, Nunes-de-Souza RL. CRF receptor type 1 (but not type 2) located within the amygdala plays a role in the modulation of anxiety in mice exposed to the elevated plus maze. Horm Behav. 2016;81:59–67. doi: 10.1016/j.yhbeh.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Contarino A, Dellu F, Koob GF, Smith GW, Lee KF, Vale W, Gold LH. Reduced anxiety-like and cognitive performance in mice lacking the corticotropin-releasing factor receptor 1. Brain Res. 1999;835:1–9. doi: 10.1016/s0006-8993(98)01158-5. [DOI] [PubMed] [Google Scholar]
- Domi E, Uhrig S, Soverchia L, Spanagel R, Hansson AC, Barbier E, Heilig M, Ciccocioppo R, Ubaldi M. Genetic Deletion of Neuronal PPARgamma Enhances the Emotional Response to Acute Stress and Exacerbates Anxiety: An Effect Reversed by Rescue of Amygdala PPARgamma Function. J Neurosci. 2016;36:12611–12623. doi: 10.1523/JNEUROSCI.4127-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulawa SC. SpringerLink (Online service), editor Novelty-Induced Hypophagia. In: Gould TD, editor. Neuromethods, Mood and Anxiety Related Phenotypes in Mice Characterization Using Behavioral Tests. 1. 2007. p. XII.p. 334.p. 352. illus. [Google Scholar]
- Gilpin NW, Herman MA, Roberto M. The central amygdala as an integrative hub for anxiety and alcohol use disorders. Biol Psychiatry. 2015;77:859–869. doi: 10.1016/j.biopsych.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Vecchiarelli HA, Morena M, Lee TT, Hermanson DJ, Kim AB, McLaughlin RJ, Hassan KI, Kuhne C, Wotjak CT, Deussing JM, Patel S, Hill MN. Corticotropin-releasing hormone drives anandamide hydrolysis in the amygdala to promote anxiety. J Neurosci. 2015;35:3879–3892. doi: 10.1523/JNEUROSCI.2737-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinks GL, Shah B, French SJ, Campos LS, Staley K, Hughes J, Sofroniew MV. Expression of LIM protein genes Lmo1, Lmo2, and Lmo3 in adult mouse hippocampus and other forebrain regions: differential regulation by seizure activity. J Neurosci. 1997;17:5549–5559. doi: 10.1523/JNEUROSCI.17-14-05549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang BH, Stewart R, Zhang JK, Lumeng L, Li TK. Corticotropin-releasing factor gene expression is down-regulated in the central nucleus of the amygdala of alcohol-preferring rats which exhibit high anxiety: a comparison between rat lines selectively bred for high and low alcohol preference. Brain Res. 2004;1026:143–150. doi: 10.1016/j.brainres.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Isogai E, Ohira M, Ozaki T, Oba S, Nakamura Y, Nakagawara A. Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS One. 2011;6:e19297. doi: 10.1371/journal.pone.0019297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Bucholz K, Dick D, Harari O, Hesselbrock V, Kramer J, Nurnberger JI, Jr, Rice J, Saccone N, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T, Goate A. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet. 2013;132:1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konnopka A, Leichsenring F, Leibing E, Konig HH. Cost-of-illness studies and cost-effectiveness analyses in anxiety disorders: a systematic review. J Affect Disord. 2009;114:14–31. doi: 10.1016/j.jad.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Kushner MG, Sher KJ, Beitman BD. The relation between alcohol problems and the anxiety disorders. Am J Psychiatry. 1990;147:685–695. doi: 10.1176/ajp.147.6.685. [DOI] [PubMed] [Google Scholar]
- Lacerda-Pinheiro SF, Pinheiro RF, Junior, Pereira de Lima MA, Lima da Silva CG, do Vieira dos Santos MS, Teixeira AG, Junior, Lima de Oliveira PN, Ribeiro KD, Rolim-Neto ML, Bianco BA. Are there depression and anxiety genetic markers and mutations? A systematic review. J Affect Disord. 2014;168:387–398. doi: 10.1016/j.jad.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Larsen S, Yokochi T, Isogai E, Nakamura Y, Ozaki T, Nakagawara A. LMO3 interacts with p53 and inhibits its transcriptional activity. Biochemical and biophysical research communications. 2010;392:252–257. doi: 10.1016/j.bbrc.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Giorgetti F, Berger KH, Tayor S, Heberlein U. Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res. 2011;35:1600–1606. doi: 10.1111/j.1530-0277.2011.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Kapfhamer D, Kharazia V, Gesch J, Giorgetti F, Heberlein U. Lmo4 in the nucleus accumbens regulates cocaine sensitivity. Genes Brain Behav. 2010;9:817–824. doi: 10.1111/j.1601-183X.2010.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos J, Husa J, Mitterer G, Haschemi A, Rauscher S, Haas R, Groger M, Loewe R, Kohrgruber N, Schrogendorfer KF, Prager G, Beck H, Pospisilik JA, Zeyda M, Stulnig TM, Patsch W, Wagner O, Esterbauer H, Bilban M. Human but not mouse adipogenesis is critically dependent on LMO3. Cell Metab. 2013;18:62–74. doi: 10.1016/j.cmet.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maiya R, Kharazia V, Lasek AW, Heberlein U. Lmo4 in the basolateral complex of the amygdala modulates fear learning. PLoS One. 2012;7:e34559. doi: 10.1371/journal.pone.0034559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Mehta RL, Molnar BE, Walters EE, Swendsen JD, Aguilar-Gaziola S, Bijl R, Borges G, Caraveo-Anduaga JJ, DeWit DJ, Kolody B, Vega WA, Wittchen HU, Kessler RC. Comorbidity of substance use disorders with mood and anxiety disorders: results of the International Consortium in Psychiatric Epidemiology. Addict Behav. 1998;23:893–907. doi: 10.1016/s0306-4603(98)00076-8. [DOI] [PubMed] [Google Scholar]
- Muller MB, Zimmermann S, Sillaber I, Hagemeyer TP, Deussing JM, Timpl P, Kormann MS, Droste SK, Kuhn R, Reul JM, Holsboer F, Wurst W. Limbic corticotropin-releasing hormone receptor 1 mediates anxiety-related behavior and hormonal adaptation to stress. Nat Neurosci. 2003;6:1100–1107. doi: 10.1038/nn1123. [DOI] [PubMed] [Google Scholar]
- Olsavszky V, Ulbrich F, Singh S, Diett M, Sticht C, Schmid CD, Zierow J, Wohlfeil SA, Schledzewski K, Dooley S, Gaitantzi H, Breitkopf-Heinlein K, Geraud C, Goerdt S, Koch PS. GATA4 and LMO3 balance angiocrine signaling and autocrine inflammatory activation by BMP2 in liver sinusoidal endothelial cells. Gene. 2017;627:491–499. doi: 10.1016/j.gene.2017.06.051. [DOI] [PubMed] [Google Scholar]
- Qin Z, Zhou X, Pandey NR, Vecchiarelli HA, Stewart CA, Zhang X, Lagace DC, Brunel JM, Beique JC, Stewart AF, Hill MN, Chen HH. Chronic stress induces anxiety via an amygdalar intracellular cascade that impairs endocannabinoid signaling. Neuron. 2015;85:1319–1331. doi: 10.1016/j.neuron.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Refojo D, Schweizer M, Kuehne C, Ehrenberg S, Thoeringer C, Vogl AM, Dedic N, Schumacher M, von Wolff G, Avrabos C, Touma C, Engblom D, Schutz G, Nave KA, Eder M, Wotjak CT, Sillaber I, Holsboer F, Wurst W, Deussing JM. Glutamatergic and dopaminergic neurons mediate anxiogenic and anxiolytic effects of CRHR1. Science. 2011;333:1903–1907. doi: 10.1126/science.1202107. [DOI] [PubMed] [Google Scholar]
- Regier DA, Narrow WE, Rae DS. The epidemiology of anxiety disorders: the Epidemiologic Catchment Area (ECA) experience. J Psychiatr Res. 1990;24(Suppl 2):3–14. doi: 10.1016/0022-3956(90)90031-k. [DOI] [PubMed] [Google Scholar]
- Sanders SK, Shekhar A. Regulation of anxiety by GABAA receptors in the rat amygdala. Pharmacol Biochem Behav. 1995;52:701–706. doi: 10.1016/0091-3057(95)00153-n. [DOI] [PubMed] [Google Scholar]
- Savarese A, Zou ME, Kharazia V, Maiya R, Lasek AW. Increased behavioral responses to ethanol in Lmo3 knockout mice. Genes Brain Behav. 2014;13:777–783. doi: 10.1111/gbb.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serretti A, Chiesa A, Calati R, Perna G, Bellodi L, De Ronchi D. Common genetic, clinical, demographic and psychosocial predictors of response to pharmacotherapy in mood and anxiety disorders. Int Clin Psychopharmacol. 2009;24:1–18. doi: 10.1097/YIC.0b013e32831db2d7. [DOI] [PubMed] [Google Scholar]
- Shekhar A, Truitt W, Rainnie D, Sajdyk T. Role of stress, corticotrophin releasing factor (CRF) and amygdala plasticity in chronic anxiety. Stress. 2005;8:209–219. doi: 10.1080/10253890500504557. [DOI] [PubMed] [Google Scholar]
- Silberman Y, Winder DG. Corticotropin releasing factor and catecholamines enhance glutamatergic neurotransmission in the lateral subdivision of the central amygdala. Neuropharmacology. 2013;70:316–323. doi: 10.1016/j.neuropharm.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GW, Aubry JM, Dellu F, Contarino A, Bilezikjian LM, Gold LH, Chen R, Marchuk Y, Hauser C, Bentley CA, Sawchenko PE, Koob GF, Vale W, Lee KF. Corticotropin releasing factor receptor 1-deficient mice display decreased anxiety, impaired stress response, and aberrant neuroendocrine development. Neuron. 1998;20:1093–1102. doi: 10.1016/s0896-6273(00)80491-2. [DOI] [PubMed] [Google Scholar]
- Sotnikov SV, Markt PO, Malik V, Chekmareva NY, Naik RR, Sah A, Singewald N, Holsboer F, Czibere L, Landgraf R. Bidirectional rescue of extreme genetic predispositions to anxiety: impact of CRH receptor 1 as epigenetic plasticity gene in the amygdala. Transl Psychiatry. 2014;4:e359. doi: 10.1038/tp.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa T, Ishikawa A, Cai LY, Kato T, Matsumoto K, Kitahara K, Kurokawa R, Ono T, Kato Y. The highly related LIM factors, LMO1, LMO3 and LMO4, play different roles in the regulation of the pituitary glycoprotein hormone alpha-subunit (alpha GSU) gene. Biosci Rep. 2009;30:51–58. doi: 10.1042/BSR20090020. [DOI] [PubMed] [Google Scholar]
- Sztainberg Y, Kuperman Y, Tsoory M, Lebow M, Chen A. The anxiolytic effect of environmental enrichment is mediated via amygdalar CRF receptor type 1. Mol Psychiatry. 2010;15:905–917. doi: 10.1038/mp.2009.151. [DOI] [PubMed] [Google Scholar]
- Timpl P, Spanagel R, Sillaber I, Kresse A, Reul JM, Stalla GK, Blanquet V, Steckler T, Holsboer F, Wurst W. Impaired stress response and reduced anxiety in mice lacking a functional corticotropin-releasing hormone receptor 1. Nat Genet. 1998;19:162–166. doi: 10.1038/520. [DOI] [PubMed] [Google Scholar]
- Tse E, Smith AJ, Hunt S, Lavenir I, Forster A, Warren AJ, Grutz G, Foroni L, Carlton MB, Colledge WH, Boehm T, Rabbitts TH. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol. 2004;24:2063–2073. doi: 10.1128/MCB.24.5.2063-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen K, Hilakivi LA, Paivarinta P, Korpi ER. Behavior of alcohol-preferring AA and alcohol-avoiding ANA rat lines in tests of anxiety and aggression. Alcohol. 1990;7:349–353. doi: 10.1016/0741-8329(90)90094-s. [DOI] [PubMed] [Google Scholar]
- Van Pett K, Viau V, Bittencourt JC, Chan RK, Li HY, Arias C, Prins GS, Perrin M, Vale W, Sawchenko PE. Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. J Comp Neurol. 2000;428:191–212. doi: 10.1002/1096-9861(20001211)428:2<191::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Zhao Y. The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein-protein interaction. Biol Cell. 2007;99:489–502. doi: 10.1042/BC20060126. [DOI] [PubMed] [Google Scholar]
- Zorrilla EP, Valdez GR, Nozulak J, Koob GF, Markou A. Effects of antalarmin, a CRF type 1 receptor antagonist, on anxiety-like behavior and motor activation in the rat. Brain Res. 2002;952:188–199. doi: 10.1016/s0006-8993(02)03189-x. [DOI] [PubMed] [Google Scholar]