Significance
We demonstrate the use of next-generation sequencing technology (RNA-seq) to understand the effect of a two-dimensional nanomaterial on human stem cells at the whole-transcriptome level. Our results identify more than 4,000 genes that are significantly affected, and several biophysical and biochemical pathways are triggered by nanoparticle treatment. We expect that this systematic approach to understand widespread changes in gene expression due to nanomaterial exposure is key to develop new bioactive materials for biomedical applications.
Keywords: 2D nanomaterials, nanosilicates, human mesenchymal stem cells, whole-transcriptome sequencing, RNA-seq
Abstract
Two-dimensional nanomaterials, an ultrathin class of materials such as graphene, nanoclays, transition metal dichalcogenides (TMDs), and transition metal oxides (TMOs), have emerged as a new generation of materials due to their unique properties relative to macroscale counterparts. However, little is known about the transcriptome dynamics following exposure to these nanomaterials. Here, we investigate the interactions of 2D nanosilicates, a layered clay, with human mesenchymal stem cells (hMSCs) at the whole-transcriptome level by high-throughput sequencing (RNA-seq). Analysis of cell–nanosilicate interactions by monitoring changes in transcriptome profile uncovered key biophysical and biochemical cellular pathways triggered by nanosilicates. A widespread alteration of genes was observed due to nanosilicate exposure as more than 4,000 genes were differentially expressed. The change in mRNA expression levels revealed clathrin-mediated endocytosis of nanosilicates. Nanosilicate attachment to the cell membrane and subsequent cellular internalization activated stress-responsive pathways such as mitogen-activated protein kinase (MAPK), which subsequently directed hMSC differentiation toward osteogenic and chondrogenic lineages. This study provides transcriptomic insight on the role of surface-mediated cellular signaling triggered by nanomaterials and enables development of nanomaterials-based therapeutics for regenerative medicine. This approach in understanding nanomaterial–cell interactions illustrates how change in transcriptomic profile can predict downstream effects following nanomaterial treatment.
Two-dimensional nanomaterials have gained unprecedented attention due to their unique atomically thin, layered, and well-defined structure that provides distinctive physical and chemical properties compared with bulk 3D counterparts (1–3). As the dimensions of 2D nanomaterials are only a few nanometers thick, they interact with biological moieties in a unique way and have raised exciting questions about their interactions with cellular components. In addition, different physical (e.g., size, shape, and charge) and chemical characteristics of 2D nanoparticles have a multitude of effects on cells including toxicity, bioactivity, or therapeutic capabilities, which are not well understood (4, 5).
Understanding cellular responses following treatment with nanomaterials will aid in evaluating their application for a range of biomedical and biotechnology applications. Recent emergence in “omics” techniques providing readouts of different biological processes, have allowed us to understand complex biological interactions of synthetic nanoparticles and their toxicity (6–9). Specifically, transcriptomics and proteomics have laid down the necessary foundation to provide an unbiased global view of the cellular activity with pivotal insights about the affected cellular pathways. Based on these results, a range of nanotechnology-based platforms have been developed for molecular diagnostics and genome-wide analysis (10). We propose to utilize transcriptomics, high-throughput sequencing of expressed transcripts (RNA-seq), to provide a holistic view of nanomaterial interactions with the cellular machinery. RNA-seq is a powerful tool for an accurate quantification of expressed transcripts that largely overcomes limitations and biases of microarrays (11–13). In this study, we will evaluate the potential of bioactive 2D nanomaterials for regenerative medicine by uncovering molecular targets and affected signaling pathways at the whole-transcriptome level.
Synthetic 2D nanoclays have been recently evaluated for regenerative medicine applications, due to their biocompatible characteristics, high surface-to-volume ratio, and uniform shape compared with other types of 2D nanomaterials (3, 14–16). Synthetic clays such as nanosilicates (Na+0.7[(Mg5.5Li0.3Si8O20(OH)4]−0.7, Laponite XLG) have disk-shaped morphology and exhibit a dual charged surface (17–19). Nanosilicates dissociate into nontoxic products [Na+, Mg2+, Si(OH)4, Li+] in physiological conditions and show 1/10th of the cytotoxicity [inhibitory concentration (IC50) ∼ 4 mg/mL] (20, 21) compared with other 2D nanomaterials such as graphene (IC50 ∼ 100 μg/mL) (22). These 2D nanosilicates are investigated for a range of biomedical applications including tissue engineering, drug and therapeutic delivery, and bioprinting (23–26). While these studies have generated encouraging results for 2D nanosilicates, their interactions affecting the transcriptome profiles remain unknown.
Here, we investigate the interactions of 2D nanosilicates with human mesenchymal stem cells (hMSCs) by employing transcriptome dynamics to uncover triggered biophysical and biochemical cellular pathways. In doing so, we observed widespread changes in gene expression profile (>4,000 genes) following nanosilicate exposure, which has not been reported previously. In addition, transcriptomic dynamics of nanosilicate-treated hMSCs identifies key genes and enriched gene ontology (GO) pathways and categories related to stem cell differentiation, specifically toward osteochondral lineages. We validated the RNA-seq findings using in vitro studies, which support the ability of nanosilicates to direct hMSC differentiation toward bone and cartilage lineages. Our study also investigated surface-mediated kinase signaling triggered by 2D nanosilicates. This work enables further development of nanomaterial-based therapeutics for regenerative medicine. More generally, transcriptomic analysis by next-generation sequencing provides a comprehensive and objective snapshot of cellular behavior following nanomaterial exposure/attachment. Furthermore, this study demonstrates the utility of next-generation sequencing for the study of cellular interactions on nanoengineered substrates and the role this approach is likely to play in this rapidly expanding field of regenerative medicine.
Biophysical and Biochemical Characterization of Nanosilicates
Chemical and structural characteristics of 2D nanomaterials will dictate their interactions with cells (5). A range of material characterization techniques was used to establish the chemical composition, crystalline nature, shape, and size of nanomaterials (27). Transmission electron microscopy (TEM) showed that nanosilicates were 20–50 nm in diameter (Fig. 1A and Fig. S1A). X-ray photoelectron spectroscopy (XPS) revealed the presence of oxygen (54.68%), silicon (28.99%), magnesium (15.27%), sodium (0.84%), and lithium (trace), which is similar to expected stoichiometry (Na+0.7[(Mg5.5Li0.3Si8O20(OH)4]−0.7) (Fig. S1B). The thickness of nanosilicates was determined to be around 1–2 nm using atomic force microscopy (AFM) (Fig. S1C). The crystalline structure of nanosilicates was corroborated by observing characteristic diffraction planes (001), (100), (005), (110), (200), and (300) using X-ray diffraction (XRD) (Fig. S1D). After exfoliation, a decrease in 2θ (20.1° to 16.8°) for diffraction plane (100), indicates an increase in d-spacing between nanosilicates.
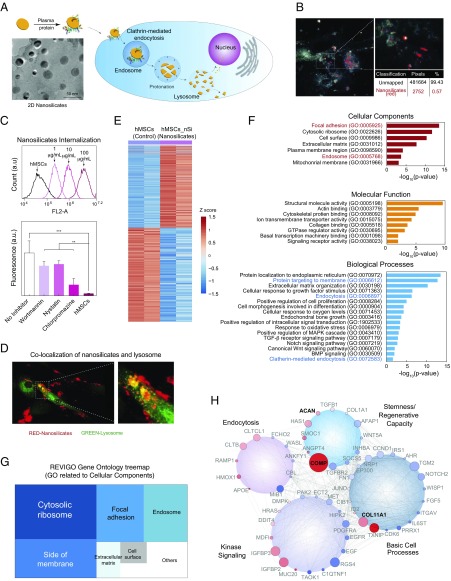
Fig. 1.
Biophysical interaction of nanosilicates and hMSCs. (A) Two-dimensional nanosilicates electrostatically bind to proteins from biological fluids and are subsequently internalized by cells via surface-mediated endocytosis. (B) Hyperspectral imaging indicating distribution of nanosilicates throughout the cell body following endocytosis. The image was captured from transverse section of cell body. (C) Flow cytometry analysis of rhodamine-tagged nanosilicates demonstrate dose-dependent cellular uptake. The nanosilicates were primarily internalized via clathrin-mediated process (chlorpromazine) as opposed to macropinocytosis (wortmannin) or caveolar-mediated (nystatin). **P < 0.01; ***P < 0.001. (D) LAMP1 staining (green) for lysosomal membranes further tracks nanosilicates (red) following endocytosis. (E) Row-scaled z-scores of quantile normalized gene expression [in log2(RPKM)] of >4,000 genes following treatment with nanosilicates (padjust < 0.05, red, up-regulated: 1,897 genes; blue, down-regulated: 2,171 genes). (F) Significant GO terms of associated biological processes, cellular components, and molecular functions from differentially regulated genes (P < 0.05). Terms related to biological process and cellular components indicate strong biophysical interactions between cells and nanosilicates. (G) Clustering of significant 244 cellular component gene ontology (GO) terms into broader cellular component categories. (H) Gene network displaying interconnected genetic targets after nanosilicate treatment with high degrees of expression and statistical significance (red, up-regulated; blue, down-regulated; size increases with significance).
In biological media, such as blood plasma, synovial fluid, or even culture media, the surface of nanoparticles becomes coated with various biomolecules forming a protein corona. Oftentimes, this initiates internalization of nanoparticles via receptor-mediated endocytosis (Fig. 1A). The binding of proteins to the nanosilicate surface was evaluated by monitoring hydrodynamic diameter (DH) and zeta potential (ζ). After mixing nanosilicates with media (containing FBS), zeta potential of the nanoparticles shifted from −40 to −25 mV, indicating that the negatively charged surface of the nanosilicates was coated with biomolecules. Similarly, an increase in hydrodynamic diameter was observed from ∼45 to ∼90 nm after placement in biological media. These results indicated that nanosilicate surfaces strongly interacted with biomolecules via electrostatic interactions to result in physical adsorption, thereby enhancing interactions at the nano–bio interface.
The effect of nanosilicates on cell health was evaluated by monitoring cytoskeletal organization, metabolic activity, and cell cycle (Fig. S2). An investigation into cell health via metabolic and viability assays [Alamar Blue and 3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT)] confirmed cytocompatibility of nanosilicates until the concentration of nanosilicates reached 100 µg/mL. In addition, hMSCs treated with nanosilicates showed similar cytoskeletal organization to untreated hMSCs. Cell cycle analysis also supported that the majority of cells were in G1 and G2 phases when treated with <100 µg/mL nanosilicates. These studies highlight that nanosilicates are cytocompatible.
Receptor-Mediated Endocytosis of Nanosilicates
The adsorbed proteins on the nanomaterial surface are predicted to influence cell surface receptor-mediated cellular uptake. We used hyperspectral imaging (28) to visualize internalized nanosilicates (Fig. 1B) without requiring chemical modifications that could have impacted uptake dynamics. Flow cytometry also demonstrated uptake of fluorescently labeled nanosilicates in a concentration-dependent manner (Fig. 1C and Fig. S3A; R2 = 0.996).
The mechanism behind nanosilicate (50 µg/mL) internalization was evaluated using chemical inhibitors to block specific endocytic pathways. We observed a significant decrease in cellular uptake of nanosilicates (79.5% reduction) when treated with a clathrin inhibitor (chlorpromazine hydrochloride) (Fig. 1D). Alternatively, other endocytic mechanisms such as caveolar-mediated (nystatin) and macropinocytosis (wortmannin) played a less prominent role in nanosilicate uptake. Furthermore, nanosilicate binding to the cell membrane and subsequent rapid internalization within 5 min (Fig. S3B) are consistent with clathrin vesicle dynamics (29). Colocalization of nanosilicates near or within lysosomal vesicles further confirmed nanosilicate trafficking (Fig. 1D). These results indicated that nanosilicates are readily internalized by cells via clathrin-mediated endocytosis and transported to degradative cell machinery.
Following uptake, nanosilicates remained within the cell for more than 7 d and were not exocytosed or dissociated immediately. The retention of nanosilicates by cells was determined using flow cytometry. A steady decrease was observed over a course of 7 d in cells staining positive for nanosilicates, that is, day 1 (96.3 ± 4.8%), day 3 (69.0 ± 10.6%), and day 7 (32.8 ± 19.5%) (Fig. S3C). To further confirm this, we monitored nanosilicate retention by cells over a week with inductively coupled plasma mass spectrometry (ICP-MS) (Fig. S3D). The nanosilicate content was decreased by 31% on day 7, compared with day 1. Over the course of 7 d, cells also maintained an enhanced lysosomal vesicle response (Fig. S3C). The stability of nanosilicates in physiological microenvironment was evaluated at pH 7.4 (mimicking cell body) and pH 5.5 (mimicking an intracellular compartment such as lysosome). A significantly higher release of minerals at pH 5.5 was observed compared with 7.4, indicating the predicted in vitro dissociation of nanosilicates. After 7 d, release of silicon (∼10%), magnesium (∼6%), and lithium (∼16%) was observed at pH 5.5. These results indicate that nanosilicates were retained by hMSCs and possibly dissociate within lysosomes over a course of 7 d.
Dissolution of nanosilicates inside cells can trigger biochemical signaling via release of minerals in the cytosol. Earlier studies have shown that mineral ions can significantly influence cell functions. For example, silicon ions have been shown to direct stem cell differentiation by triggering cWnt signaling pathways and are critical for cartilage development (30, 31). Likewise, magnesium ions have been shown to up-regulate production of COL10A1 and VEGF in hMSCs (32). Lithium, an inhibitor of glycogen synthetase kinase-3β (GSK-3β), activates Wnt-responsive genes by elevating cytoplasmic β-catenin (33, 34). These studies suggest that intracellular release of ionic dissolution products of nanosilicates [Si(OH)4, Mg2+, Li+] could stimulate hMSC differentiation.
Widespread Transcriptomic Changes Triggered by Nanosilicates
Sequencing of expressed mRNAs by RNA-seq can be used to determine genome-wide changes in gene expression resulting from cellular response to external stimuli (35). hMSCs (2,500 cells per cm2) were exposed to nanosilicates (50 µg/mL), and whole-transcriptome sequencing (RNA-seq) was performed after 7 d (Materials and Methods). The 7-d time point was chosen to provide a broad overview of cell processes, ranging from endocytosis and proliferation to early differentiation. Two replicates of untreated and treated hMSCs were sequenced. The sequenced reads were aligned to reference genome (hg19) using RNA-seq aligner. The normalized gene expression levels were determined by calculating the reads per kilobase of transcript per million (RPKM) (Fig. S4 A and B). The replicates for both the conditions showed high concordance (r = 0.99, Fig. S4C). We used generalized linear models (GLMs) to identify differential gene expression (DGE) between nanosilicate treated hMSCs and untreated hMSCs (SI Materials and Methods). This comparison revealed significant changes in the expression level of 4,068 genes (Fig. 1E and Dataset S1; 1,897 up-regulated genes, 2,171 down-regulated genes, false-discovery rate-adjusted P < 0.05). Such widespread changes in gene expression profile have not been reported earlier. For example, human dermal fibroblast cells treated with gold nanoparticles were shown to differentially regulate 1,439 genes (36), while another study demonstrated that human immune cells treated with graphene oxide experienced differential regulation of 1,147 genes (37). It is important to note that these previously reported studies were performed using microarrays (36, 37) and the widespread effect of nanoparticles on whole transcriptome was not investigated. Thus, our result strongly suggests that nanoparticle treatment leads to a widespread cellular response that is reflected by the change in transcriptome profile of hMSCs treated with nanosilicates, requiring further exploration into prominent cellular pathways.
DGE following nanosilicate introduction spanned a host of cellular processes and functions. To identify the key biological processes and pathways that are affected when the cells interact with nanosilicates, we performed GO enrichment analysis for the three GO categories [biological processes (BP), cellular components (CC), and molecular functions (MF)]. Nanosilicate treatment showed significant enrichment for 1,132 GO terms (P < 0.05), including 884 for BP, 134 for CC, and 114 for MF (Fig. S4D and Dataset S2). We then narrowed down key GO terms based on high significance (P value) in each category to highlight the widespread effect of nanosilicates on hMSCs (Fig. 1F). The key GO terms significantly enriched in BP were endocytosis (GO:0006897) and endochondral bone growth (GO:0003416). The analysis also indicated positive regulation of mitogen-activated protein kinase (MAPK) cascade (GO:0043410), transforming growth factor-β (TGF-β) receptor signaling pathway (GO:0007179), notch signaling pathway (GO:0007219), canonical Wingless (cWnt) signaling pathway (GO:0060070), and bone morphogenic protein (BMP) signaling (GO:0030509). GO analysis also supported our observation that nanosilicates are internalized via clathrin-mediated endocytosis (GO:0072583). Overall, the GO enrichment analysis indicated that the predominant downstream effect of nanosilicates was on kinase activity, cell differentiation, and extracellular matrix (ECM) reorganization.
Functional annotation clustering performed using Database for Annotation Visualization and Integrated Discovery (DAVID) (38) highlights the role of cell membrane-mediated signaling due to nanosilicate treatment (Dataset S3). We then used REVIGO (39) to refine the extensive list of significant CC GO terms by reducing functional redundancies and clustering the terms based on semantic similarity measures. GO for CC was enriched for cytosolic, ribosome, focal adhesion, and endosomal processes (Fig. 1G and Fig. S4E). These results further suggested a sequence of events initiated at the cell membrane through protein localization to membrane (GO:0072657) and endocytic vesicle formation (GO:0006897, GO:0006898) accompanied by protein targeting to membrane (GO:0006612), and trafficking by lysosome (GO:0043202, GO:0005764). Specifically, genes involved in clathrin-mediated endocytosis (GO:0072583) like CLTCL1, which encodes a major protein of the polyhedral pit and vesicle coat, were significantly affected (Dataset S1). To validate the clathrin-mediated endocytosis, change in expression level of CLTCL1 was confirmed using quantitative reverse transcription–PCR (qRT-PCR) (Fig. S4F). We then clustered differentially expressed genes (P < 0.05) using Cytoscape (40) into different cellular processes such as basic cell processes, kinase signaling, endocytosis, and stemness/regenerative capacity (SI Materials and Methods). Networks between genes from same pathways were generated to illustrate connected and interdependent genes regulated by nanosilicate treatment (Fig. 1H and Fig. S5). Overall, these results demonstrate that nanosilicates significantly affect the transcriptomic profile of hMSCs, which can translate to measurable changes in behavior.
Nanosilicates Activate Surface-Mediated Signaling
The high surface-to-volume ratio and dual charged surface of nanosilicates are expected to facilitate strong interactions with the cell membrane. The physical interactions between cells and nanoparticles are expected to stimulate a variety of intracellular signaling events including proliferation and differentiation (5, 41, 42). Accordingly, a significant change in expression of upstream regulators of Ras (e.g., RalB, DDIT4, and HRAS) and Rho (e.g., DMPK, PAK2, and ECT2) subfamilies of GTPases was observed upon nanosilicate treatment (Dataset S1). These Ras and Rho genes are associated with peptidyl-serine phosphorylation (GO:0033135) and protein serine/threonine kinase activity (GO:0071900). Both Ras and Rho GTPase subfamilies affect cell behaviors such as cytoskeletal arrangement, cell migration, and stem cell fate (43, 44). From analyzing enriched GO pathways related to stress, two prominent membrane-activated cascades emerged: the MAPK cascade and Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway (Fig. 2A). Genetic markers specific to extracellular signal-regulated kinases (ERKs) ERK1/ERK2 regulation (GO:0070374, GO:0071363), stress-activated MAPK (GO:0032872, GO:0031098) cascades, and JAK/STAT cascade (GO:0007259) were also significantly altered following nanosilicate treatment. Among these enriched GO terms, multiple genes displayed notable log2fold changes in expression such as IGFBP2 (insulin-like growth factor binding protein 2) (log2fold: 1.358), IGFBP3 (1.149), TAOK1 (−1.864), PDGFRA (−1.394), and HIPK2(−1.237). A significant change in gene expression of key MAPK signaling regulators was observed (Fig. S6B).
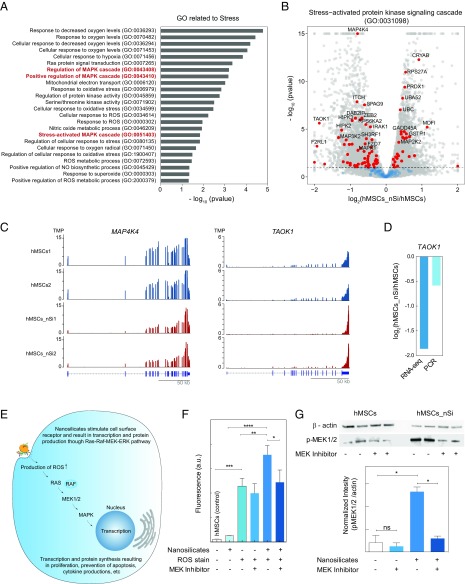
Fig. 2.
Nanosilicates lead to stress-induced MAPK signaling. (A) Nanosilicate treatment results in activation of stress-related response. A list of significant GO terms related to stress after nanosilicate treatment indicate signal propagation via MAPK/ERK signaling pathways. (B) The majority of genes involved in stress-activated kinase signaling cascade (GO:0031098) undergo a significant differential expression. (C) The change in gene expression profile of MAP4K4 and TAOK1 (aligned reads normalized by total library size). (D) Comparison of TAOK1 gene expression obtained from RNA-seq was validated using qRT-PCR. (E) Nanosilicates trigger a stress-responsive kinase cascade (Ras–Raf–MEK–ERK pathways), leading to changes in reactive oxygen species (ROS) production and subsequent RNA transcription and protein synthesis. (F) Flow-cytometric analysis was performed to measure the stress-responsive kinase cascade, by measuring ROS production with a ROS-sensitive fluorescent reporter dye. Experiments were performed in the presence or absence of a MAPK inhibitor. A significant increase in ROS-mediated fluorescent signal is observed upon exposure to nanosilicate, and this is abrogated after treatment with the MAPK inhibitor. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (G) Production of p-MEK1/2 was determined using Western blot in presence of nanosilicates and MEK inhibitor, establishing the role of nanosilicate in MAPK/ERK signaling. *P < 0.05. ns, not significant.
We also observed that a large number of genes (76 out of 170 genes) related to stress-activated protein kinase signaling (GO:0031098) were differentially expressed due to nanosilicates treatment (Fig. 2B). Specifically, TAOK1, TXNIP, and MAP4K4 exhibited a distinct difference in expression between nanosilicate-treated hMSCs compared with control hMSCs (Fig. 2C and Fig. S6C). TAOK1 is an activator of the p38/MAPK14 stress-activated MAPK cascade (45). The change in mRNA expression levels of TAOK1 via RNA-seq was further validated using qRT-PCR (Fig. 2D). These data strongly support the ability of nanosilicates to stimulate MAPK cascade, specifically that of the ERK and p38 pathways (Fig. 2E).
To experimentally validate the cross talk between MAPK signaling pathways following nanosilicate treatment, flow-cytometric analysis was performed. Reactive oxygen species (ROS) produced by cells treated with and without nanosilicates in presence of ERK inhibitor [PD184352, mitogen-activated protein kinase 1/2 (MEK1/2) inhibitor] were monitored using a ROS-sensitive reporter fluorophore (Fig. 2F). ROS play a role in the ERK pathway via cross talk from mitogen-activated protein kinase kinase kinases (46). For nanosilicate-treated hMSCs, a significant reduction in ROS production was observed due to the presence of ERK inhibitor as seen by reduced fluorescence signal (∼32% reduction, P < 0.05). This reduction in ROS production via the ERK inhibitor indicates the stimulation of MAPK signaling, specifically that of ERK, by the nanosilicates. The mechanism of this activation may stem from the biophysical cell–nanoparticle interaction, biochemical dissolution, or both in conjunction. This study validates that hMSCs recognize and respond to nanosilicates by engaging intracellular programs such as MAPK cascade (ERK1/2 and p38 kinases).
While RNA-seq analysis provided insight about the role of nanosilicate in stimulating MAPK-related pathways, including those typically stimulated by growth factors in addition to stress-responsive kinases, monitoring protein levels can further provide functional evidence. Differential expression was observed in both upstream (e.g., Ras, PRKCA, and BRAF) and downstream (e.g., ELK1, MKNK2) genes of MEK1/2. In the MAPK/ERK cascade, MEK1 and MEK2 control cell growth and differentiation (47). Activation of MEK1 and MEK2 occurs through phosphorylation by Raf. MEK1/2 inhibitors have been used extensively to implicate ERK1/2 in a wide array of biological events. To validate MAPK/ERK pathways, we monitored synthesis and phosphorylation of MEK1/2 (p-MEK1/2) with and without a MEK1/2 inhibitor (Fig. 2G) via Western blot. hMSCs had relatively low production of p-MEK1/2, while nanosilicate treatment results in more than sixfold increase in p-MEK1/2. In the presence of MEK1/2 inhibitor, production of p-MEK1/2 in the presence of nanosilicates was suppressed, demonstrating the role of nanosilicates in activating the MAPK/ERK pathway.
Beyond intracellular phosphorylation events within MAPK cascades, we also observed genes that play a role in controlling background processes of hMSCs, like multipotency and motility, that have been identified in previous literature (48). RNA-seq analysis revealed a significant change in gene expression: AFAP1 (log2foldΔ: −1.152), SOCS5 (−1.192), WNT5A (−1.162), INHBA (−1.179) from a variety of pathways including TGF-β, JAK/STAT, Wnt/β-catenin, and phosphatidylinositol-3-kinase (PI3K) signaling. As this subset of genes is involved in cell proliferation, stromal cell multipotency, and extracellular matrix production (49–51), nanosilicates may therefore improve functional tissue regeneration. Therefore, we were prompted to investigate these downstream pathways using molecular analysis techniques.
Nanosilicates Direct Stem Cell Differentiation
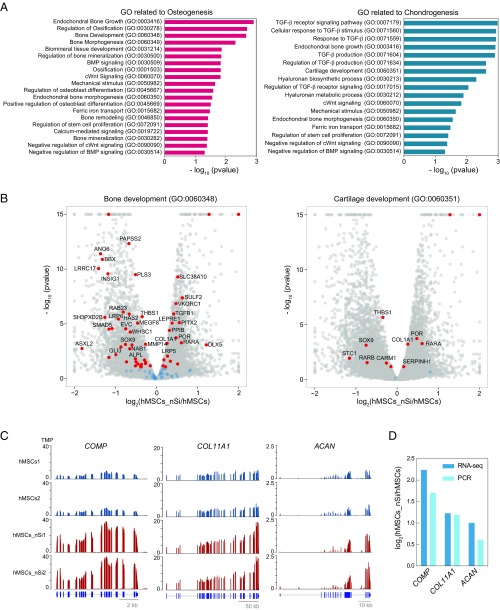
Following nanosilicates treatment, activation of the membrane can lead to differentiation and extracellular matrix deposition, following an ERK-based cascade. The kinase signaling follows similar progressions in hMSCs after growth factor stimulation to promote osteochondral differentiation (52–56). Evidence of hMSC inclination toward bone and cartilage lineages following nanosilicate treatment was observed with GO term enrichment (Fig. 3A and Fig. S7). GO pathways and biological processes related to osteogenesis, such as bone development (GO:0060348), endochondral bone growth (GO:0003416), biomineral tissue development (GO:0031214), and canonical Wnt signaling pathway (GO:0060070) were favored toward osteogenesis. For chondrogenesis, cellular response to transforming growth factor-β stimulus (GO:0071560), cartilage development involved in endochondral bone morphogenesis (GO:0060351), and hyaluronan metabolic process (GO:0030212) were significantly altered. We observed a large fraction of genes with differential expression due to nanosilicate treatment. For example, 49 out of 92 genes were differentially expressed for bone development (GO:0060348), while 10 out of 14 genes were differentially expressed for cartilage development (GO:0060351) (Fig. 3B). Genes from these GO categories including cartilage oligomeric matrix protein (COMP), collagen type I α1 chain (COL1A1), collagen type XI α1 chain (COL11A1), and aggrecan (ACAN), were significantly up-regulated due to nanosilicate treatment (Fig. 3C and Fig. S8A). We further validated these genes using qRT-PCR and observed comparable gene expression to that of RNA-seq (Fig. 3D).
Fig. 3.
Transcriptomic analysis elucidates nanosilicate-induced bioactivity. (A) GO terms related to osteogenesis and chondrogenesis indicate nanosilicate-induced hMSC differentiation. (B) Significant gene expression changes in genes involved in bone development (GO:0060348) and cartilage development (GO:0060351). (C) Gene expression profile of COMP, COL11A1, and ACAN, demonstrating up-regulation due to nanosilicate treatment (aligned reads normalized by total library size). (D) Differential gene expression from RNA-seq was validated using qRT-PCR, indicating similar trend.
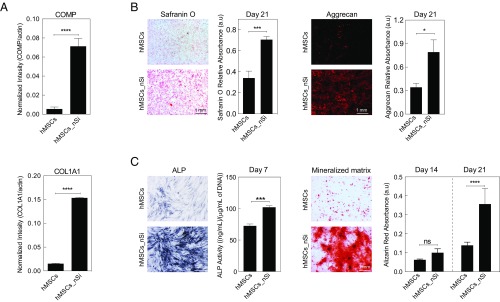
To ensure that the mRNA detected represented up-regulated protein levels, a Western blot for COMP and COL1A1 was performed on day 7. Both COMP and COL1A1 protein showed a significant increase in expression due to nanosilicate treatment, indicating their role in hMSC differentiation (Fig. 4A). Based on the changes in transcriptomic profile and in vitro validation, we hypothesize that the activation of MAPK/ERK pathways by nanosilicates may lead to differentiation into osteochondral lineages. To confirm the role nanosilicate in stimulating MAPK/ERK signaling for hMSC differentiation, a MEK1/2 inhibitor was utilized and resulted in a significant decrease in COMP protein synthesis (Fig. S8B). This indicated the role of nanosilicates in the activation of MAPK/ERK signaling to direct the differentiation of hMSCs.
Fig. 4.
Nanosilicate-induced hMSCs differentiation. (A) Western blot showing production of COL1A1 and COMP after exposure to nanosilicates for 7 d in normal media. (B) The effect of nanosilicates on production of GAGs was determined by safranin O and aggrecan staining after culturing hMSCs in chondro-conductive media for 21 d. (C) The effect of nanosilicates on osteogenic differentiation was determined by ALP activity and formation of mineralized matrix after culturing hMSCs in osteo-conductive media for 21 d. *P < 0.05; ***P < 0.001; ****P < 0.0001. ns, not significant.
Finally, to further substantiate the ability of nanosilicates to drive hMSC differentiation toward bone and cartilage lineages, staining of lineage-specific proteins and matrix mineralization was performed. The effect of nanosilicates on chondrogenic and osteogenic differentiation was monitored by subjecting nanosilicate-treated hMSCs to chondro-conductive (lacking TGF-β) and osteo-conductive [lacking bone morphogenetic protein 2 (BMP2) or dexamethasone] media. After 21 d, production of chondro- and osteo-related ECM was observed even in the absence of inductive supplements. An increase in both glycosaminoglycan (GAGs) and aggrecan production were observed in nanosilicate-treated hMSCs indicating chondrogenic differentiation (Fig. 4B), while for osteogenic differentiation, an increase in alkaline phosphatase (ALP) production as well as matrix mineralization (calcium phosphates) were observed after nanosilicate treatment (Fig. 4C). These results validate the ability of nanosilicates to induce hMSC differentiation into the bone and cartilage lineages without the use of inductive agents and growth factors.
The broader relevance of our study is that “omics” techniques can be used to determine the effect of nanomaterials on cells in a nontargeted and nonbiased approach. The physicochemical properties of engineered nanomaterials such as size, shape, surface charge, and chemical composition will have profound effects on cellular behavior (5, 41). Therefore, we do not speculate that the transcriptomic changes observed in this study will be universal to all type of nanomaterials or even single-cell analyses. Recent studies have used bulk population “omics” approaches to understand nanotoxicology and mechanism-based risk assessment of nanomaterials (57, 58). The current study demonstrates the capabilities of next-generation sequencing to monitor mRNA expression levels in the cell after nanomaterial treatment. Our approach overcomes the limitation of measuring expression levels of preselected genes on microarrays, which can therefore identify previously neglected cellular signaling pathways relevant for regeneration. Additionally, RNA-seq delivers a low background signal and sequenced reads that can be unambiguously mapped to unique regions of the genome, which will help in sensitive and precise identification of the expressed genes. The transcriptomic insight on the role of surface-mediated cellular signaling supports the ability of nanosilicates to induce hMSC differentiation into bone and cartilage lineages in the absence of inductive agents. This insight can assist in reducing or eliminating the use of supraphysiological doses of growth factors currently employed in clinical practice for regenerative therapies. These omics techniques can likewise reveal cell–material interactions unique to specific nanoparticles and can contribute directly to the design of bioactive nanomaterials for regenerative medicine.
Conclusion
Overall, we investigated a transcriptomic snapshot of hMSCs in which a widespread change in transcriptomic profile was observed in response to nanosilicate exposure. The transcriptomic changes observed due to nanosilicate treatment likely originate from both biophysical and biochemical mechanisms. The interaction of nanosilicates with the cell membrane stimulated various surface receptors, including the stress-responsive and surface receptor-mediated MAPK pathways. Similarly, the data indicate that internalization of nanosilicates and subsequent release of mineral ions trigger biochemical signaling that could promote osteochondral differentiation of hMSCs. Accordingly, analysis of the transcriptomic snapshot of hMSCs treated with nanosilicates uncovered families of genes related to osteochondral differentiation. In vitro studies validated the RNA-seq findings and further supported the observation that nanosilicates have the capacity to direct hMSC differentiation toward bone and cartilage lineages. Last, RNA-seq emerged as a viable technique to evaluate the regenerative potential of novel nanomaterials.
Materials and Methods
See Supporting Information for detailed materials and methods. Detailed methods for nanoparticle characterization, in vitro studies, and whole-transcriptome sequencing can be found in SI Materials and Methods.
Nanoparticle Characterization.
Synthetic clay nanosilicates (Laponite XLG, Na+0.7[(Mg5.5Li0.3Si8O20(OH)4]−0.7) were obtained from BYK Additives. Structural, physical, and chemical properties of nanosilicates were characterized via ICP-MS (elemental analysis; NexION 300D; PerkinElmer) and XPS (Omicron XPS system with Argus detector), XRD (Bruker D8 Advanced), AFM (Bruker Dimension Icon Nanoscope), and TEM (JEOL JEM-2010).
In Vitro Analysis.
hMSCs, obtained from Texas A&M Institute for Regenerative Medicine, were cultured under basal media conditions and used for all experiments. Nanosilicate trafficking and cytocompatibility were monitored using standard assays. For gene and protein expression, hMSCs were treated with and without nanosilicates (50 µg/mL) for 48 h; nanosilicates were then removed and cells were cultured under normal conditions for an additional 5 d. qRT-PCR and Western blot analysis were performed with standard protocols to validate gene expression and corresponding protein expression. For differentiation studies, hMSCs were treated with either osteogenic (normal media supplemented with 10 mM β-glycerophosphate and 50 µM ascorbic acid) or chondrogenic media (DMEM supplemented with 1% ITS+, 10−7 M dexamethasone, and 1 mM sodium pyruvate) with and without nanosilicates.
Whole-Transcriptome Sequencing.
hMSCs were treated with and without nanosilicates (50 µg/mL) for 48 h; nanosilicates were then removed and cells were cultured under normal conditions for an additional 5 d. Samples were analyzed via a high-output HiSeq platform with TruSeqRNA sample preparation and single-end read length of 125 bases. The sequenced reads were trimmed and aligned to the human genome (hg19). GLMs were used to identify the differentially expressed genes where the expression counts were modeled as negative binomial distribution. Only genes with a P adjusted value (false-discovery rate) of less than 0.05 were included within the network and subsequent GO term network formation. Data were analyzed using Cytoscape, GeneMANIA, ClueGO, and DAVID bioinformatics resources.
Statistical Analysis.
For in vitro studies, significance was determined via one-way ANOVA with post hoc Tukey tests. For statistical analysis of RNA-seq data, see Supporting Information.
Supplementary Material
Acknowledgments
We acknowledge Drs. Wilson Serem, Jing Wu, and Joseph Reibenspies from Texas A&M University for their assistance with AFM, XPS, and XRD characterization, respectively. We thank Dr. Charlie Johnson, Director of Genomics and Bioinformatics at Texas A&M AgriLife, for his assistance with the RNA-seq experiment. We also thank CytoViva for complimentary hyperspectral imaging data. A.K.G. acknowledges financial support from the National Institutes of Health (Grant R03 EB023454). J.K.C. acknowledges financial support from Sigma Xi Grant-in-Aid-of-Research.
Footnotes
Conflict of interest statement: J.K.C. and A.K.G. are coauthors on US Patent Application No. WO2017112802 A1 published on June 29, 2017 (US Provisional Patent Application No. 62/270,403 filed on December 21, 2015).
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE108638).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716164115/-/DCSupplemental.
References
- 1.Butler SZ, et al. Progress, challenges, and opportunities in two-dimensional materials beyond graphene. ACS Nano. 2013;7:2898–2926. doi: 10.1021/nn400280c. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Tan C, Zhang H, Wang L. Two-dimensional graphene analogues for biomedical applications. Chem Soc Rev. 2015;44:2681–2701. doi: 10.1039/c4cs00300d. [DOI] [PubMed] [Google Scholar]
- 3.Chimene D, Alge DL, Gaharwar AK. Two-dimensional nanomaterials for biomedical applications: Emerging trends and future prospects. Adv Mater. 2015;27:7261–7284. doi: 10.1002/adma.201502422. [DOI] [PubMed] [Google Scholar]
- 4.Jiang W, Kim BYS, Rutka JT, Chan WCW. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 5.Kerativitayanan P, Carrow JK, Gaharwar AK. Nanomaterials for engineering stem cell responses. Adv Healthc Mater. 2015;4:1600–1627. doi: 10.1002/adhm.201500272. [DOI] [PubMed] [Google Scholar]
- 6.Verano-Braga T, et al. Insights into the cellular response triggered by silver nanoparticles using quantitative proteomics. ACS Nano. 2014;8:2161–2175. doi: 10.1021/nn4050744. [DOI] [PubMed] [Google Scholar]
- 7.Ge Y, Bruno M, Wallace K, Winnik W, Prasad RY. Proteome profiling reveals potential toxicity and detoxification pathways following exposure of BEAS-2B cells to engineered nanoparticle titanium dioxide. Proteomics. 2011;11:2406–2422. doi: 10.1002/pmic.201000741. [DOI] [PubMed] [Google Scholar]
- 8.Simon DF, et al. Transcriptome sequencing (RNA-seq) analysis of the effects of metal nanoparticle exposure on the transcriptome of Chlamydomonas reinhardtii. Appl Environ Microbiol. 2013;79:4774–4785. doi: 10.1128/AEM.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucafò M, et al. Profiling the molecular mechanism of fullerene cytotoxicity on tumor cells by RNA-seq. Toxicology. 2013;314:183–192. doi: 10.1016/j.tox.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Larguinho M, Baptista PV. Gold and silver nanoparticles for clinical diagnostics—from genomics to proteomics. J Proteomics. 2012;75:2811–2823. doi: 10.1016/j.jprot.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 12.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 13.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Dawson JI, Oreffo RO. Clay: New opportunities for tissue regeneration and biomaterial design. Adv Mater. 2013;25:4069–4086. doi: 10.1002/adma.201301034. [DOI] [PubMed] [Google Scholar]
- 15.Ghadiri M, Chrzanowski W, Rohanizadeh R. Biomedical applications of cationic clay minerals. RSC Adv. 2015;5:29467–29481. [Google Scholar]
- 16.Viseras C, Cerezo P, Sanchez R, Salcedo I, Aguzzi C. Current challenges in clay minerals for drug delivery. Appl Clay Sci. 2010;48:291–295. [Google Scholar]
- 17.Ruzicka B, et al. Observation of empty liquids and equilibrium gels in a colloidal clay. Nat Mater. 2011;10:56–60. doi: 10.1038/nmat2921. [DOI] [PubMed] [Google Scholar]
- 18.Thompson DW, Butterworth JT. The nature of laponite and its aqueous dispersions. J Colloid Interface Sci. 1992;151:236–243. [Google Scholar]
- 19.Tawari SL, Koch DL, Cohen C. Electrical double-layer effects on the Brownian diffusivity and aggregation rate of laponite clay particles. J Colloid Interface Sci. 2001;240:54–66. doi: 10.1006/jcis.2001.7646. [DOI] [PubMed] [Google Scholar]
- 20.Gaharwar AK, et al. Bioactive silicate nanoplatelets for osteogenic differentiation of human mesenchymal stem cells. Adv Mater. 2013;25:3329–3336. doi: 10.1002/adma.201300584. [DOI] [PubMed] [Google Scholar]
- 21.Mihaila SM, et al. The osteogenic differentiation of SSEA-4 sub-population of human adipose derived stem cells using silicate nanoplatelets. Biomaterials. 2014;35:9087–9099. doi: 10.1016/j.biomaterials.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 22.Akhavan O, Ghaderi E, Akhavan A. Size-dependent genotoxicity of graphene nanoplatelets in human stem cells. Biomaterials. 2012;33:8017–8025. doi: 10.1016/j.biomaterials.2012.07.040. [DOI] [PubMed] [Google Scholar]
- 23.Xavier JR, et al. Bioactive nanoengineered hydrogels for bone tissue engineering: A growth-factor-free approach. ACS Nano. 2015;9:3109–3118. doi: 10.1021/nn507488s. [DOI] [PubMed] [Google Scholar]
- 24.Gaharwar AK, Schexnailder PJ, Kline BP, Schmidt G. Assessment of using laponite cross-linked poly(ethylene oxide) for controlled cell adhesion and mineralization. Acta Biomater. 2011;7:568–577. doi: 10.1016/j.actbio.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Wilson SA, Cross LM, Peak CW, Gaharwar AK. Shear-thinning and thermo-reversible nanoengineered inks for 3D bioprinting. ACS Appl Mater Interfaces. 2017;9:43449–43458. doi: 10.1021/acsami.7b13602. [DOI] [PubMed] [Google Scholar]
- 26.Dawson JI, Kanczler JM, Yang XB, Attard GS, Oreffo RO. Clay gels for the delivery of regenerative microenvironments. Adv Mater. 2011;23:3304–3308. doi: 10.1002/adma.201100968. [DOI] [PubMed] [Google Scholar]
- 27.Hühn J, et al. Selected standard protocols for the synthesis, phase transfer, and characterization of inorganic colloidal nanoparticles. Chem Mater. 2016;29:399–461. [Google Scholar]
- 28.Peña Mdel P, et al. Hyperspectral imaging of nanoparticles in biological samples: Simultaneous visualization and elemental identification. Microsc Res Tech. 2016;79:349–358. doi: 10.1002/jemt.22637. [DOI] [PubMed] [Google Scholar]
- 29.Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han P, Wu C, Xiao Y. The effect of silicate ions on proliferation, osteogenic differentiation and cell signalling pathways (WNT and SHH) of bone marrow stromal cells. Biomater Sci. 2013;1:379–392. doi: 10.1039/c2bm00108j. [DOI] [PubMed] [Google Scholar]
- 31.Reffitt DM, et al. Orthosilicic acid stimulates collagen type 1 synthesis and osteoblastic differentiation in human osteoblast-like cells in vitro. Bone. 2003;32:127–135. doi: 10.1016/s8756-3282(02)00950-x. [DOI] [PubMed] [Google Scholar]
- 32.Yoshizawa S, Brown A, Barchowsky A, Sfeir C. Magnesium ion stimulation of bone marrow stromal cells enhances osteogenic activity, simulating the effect of magnesium alloy degradation. Acta Biomater. 2014;10:2834–2842. doi: 10.1016/j.actbio.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Clément-Lacroix P, et al. Lrp5-independent activation of Wnt signaling by lithium chloride increases bone formation and bone mass in mice. Proc Natl Acad Sci USA. 2005;102:17406–17411. doi: 10.1073/pnas.0505259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang F, Phiel CJ, Spece L, Gurvich N, Klein PS. Inhibitory phosphorylation of glycogen synthase kinase-3 (GSK-3) in response to lithium. Evidence for autoregulation of GSK-3. J Biol Chem. 2003;278:33067–33077. doi: 10.1074/jbc.M212635200. [DOI] [PubMed] [Google Scholar]
- 35.Nel AE, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 36.Grzincic EM, Yang JA, Drnevich J, Falagan-Lotsch P, Murphy CJ. Global transcriptomic analysis of model human cell lines exposed to surface-modified gold nanoparticles: The effect of surface chemistry. Nanoscale. 2015;7:1349–1362. doi: 10.1039/c4nr05166a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orecchioni M, et al. Molecular and genomic impact of large and small lateral dimension graphene oxide sheets on human immune cells from healthy donors. Adv Healthc Mater. 2016;5:276–287. doi: 10.1002/adhm.201500606. [DOI] [PubMed] [Google Scholar]
- 38.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albanese A, Tang PS, Chan WC. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 42.Rauch J, Kolch W, Laurent S, Mahmoudi M. Big signals from small particles: Regulation of cell signaling pathways by nanoparticles. Chem Rev. 2013;113:3391–3406. doi: 10.1021/cr3002627. [DOI] [PubMed] [Google Scholar]
- 43.Liu ZJ, Zhuge Y, Velazquez OC. Trafficking and differentiation of mesenchymal stem cells. J Cell Biochem. 2009;106:984–991. doi: 10.1002/jcb.22091. [DOI] [PubMed] [Google Scholar]
- 44.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 45.Hutchison M, Berman KS, Cobb MH. Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J Biol Chem. 1998;273:28625–28632. doi: 10.1074/jbc.273.44.28625. [DOI] [PubMed] [Google Scholar]
- 46.Wang FS, et al. Superoxide mediates shock wave induction of ERK-dependent osteogenic transcription factor (CBFA1) and mesenchymal cell differentiation toward osteoprogenitors. J Biol Chem. 2002;277:10931–10937. doi: 10.1074/jbc.M104587200. [DOI] [PubMed] [Google Scholar]
- 47.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song L, Webb NE, Song Y, Tuan RS. Identification and functional analysis of candidate genes regulating mesenchymal stem cell self-renewal and multipotency. Stem Cells. 2006;24:1707–1718. doi: 10.1634/stemcells.2005-0604. [DOI] [PubMed] [Google Scholar]
- 49.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 50.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 51.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 52.Hu X, et al. The role of ERK and JNK signaling in connective tissue growth factor induced extracellular matrix protein production and scar formation. Arch Dermatol Res. 2013;305:433–445. doi: 10.1007/s00403-013-1334-9. [DOI] [PubMed] [Google Scholar]
- 53.Caro-Gonzalez HY, et al. Mitogen-activated protein kinase (MAPK/ERK) regulates adenomatous polyposis coli during growth-factor-induced cell extension. J Cell Sci. 2012;125:1247–1258. doi: 10.1242/jcs.095166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 55.Mansour SJ, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 56.Yan L, Luo H, Gao X, Liu K, Zhang Y. Vascular endothelial growth factor-induced expression of its receptors and activation of the MAPK signaling pathway during ovine oocyte maturation in vitro. Theriogenology. 2012;78:1350–1360. doi: 10.1016/j.theriogenology.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Costa PM, Fadeel B. Emerging systems biology approaches in nanotoxicology: Towards a mechanism-based understanding of nanomaterial hazard and risk. Toxicol Appl Pharmacol. 2016;299:101–111. doi: 10.1016/j.taap.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 58.Autefage H, et al. Sparse feature selection methods identify unexpected global cellular response to strontium-containing materials. Proc Natl Acad Sci USA. 2015;112:4280–4285. doi: 10.1073/pnas.1419799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.