Fig. 2.
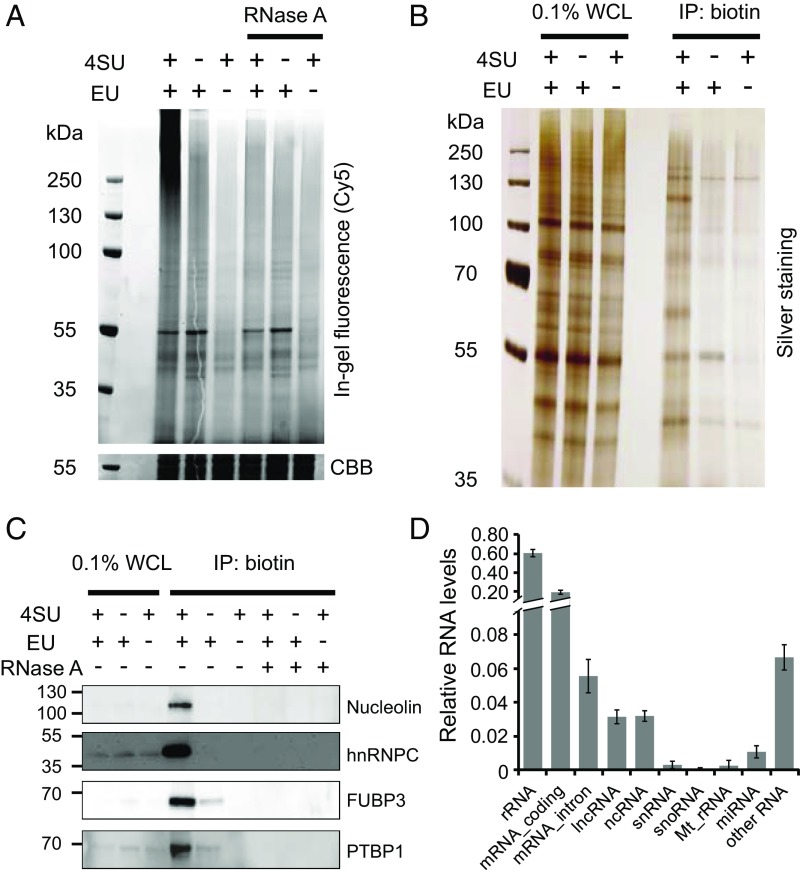
Development and optimization of CARIC. (A) In-gel fluorescence analysis of RNPs. HeLa cells were treated with 1 mM EU and 0.5 mM 4SU, EU, or 4SU alone and irradiated with 365-nm UV light. The lysates were reacted with azide-Cy5 via THPTA-assisted CuAAC. After treatment with or without RNase A, the samples were resolved by SDS/PAGE and visualized by in-gel fluorescence scanning. Coomassie brilliant blue (CBB)-stained gel was used as the loading control. (B) After EU&4SU labeling and UV light irradiation, the cell lysates were reacted with azide-biotin and enriched with streptavidin beads. After elution, the captured RNPs were digested with RNase A to release RBPs, which were resolved by SDS/PAGE and visualized by silver staining. WCL, whole-cell lysate. (C) Western blot analysis of the presence of nucleolin, hnRNPC, FUBP3, and PTBP1 in the CARIC-captured samples. RNase A treatment after click labeling of the lysates with azide-biotin demonstrated selective capture of RNPs doubly labeled with EU and 4SU. (D) Analysis of RNAs isolated by CARIC by next-generation sequencing. The relative RNA level was quantified by normalizing the total mapped reads of each RNA types to the total mapped reads of total RNAs. Error bars represent the SD from three independent biological replicates.