Abstract
Background
Neuropathological findings support an autoimmune etiology as an underlying factor for loss of orexin-producing neurons in spontaneous narcolepsy type 1 (narcolepsy with cataplexy; sNT1) as well as in Pandemrix influenza vaccine-induced narcolepsy type 1 (Pdmx-NT1). The precise molecular target or antigens for the immune response have, however, remained elusive.
Methods
Here we have performed a comprehensive antigenic repertoire analysis of sera using the next-generation phage display method - mimotope variation analysis (MVA). Samples from 64 children and adolescents were analyzed: 10 with Pdmx-NT1, 6 with sNT1, 16 Pandemrix-vaccinated, 16 H1N1 infected, and 16 unvaccinated healthy individuals. The diagnosis of NT1 was defined by the American Academy of Sleep Medicine international criteria of sleep disorders v3.
Findings
Our data showed that although the immunoprofiles toward vaccination were generally similar in study groups, there were also striking differences in immunoprofiles between sNT1 and Pdmx-NT1 groups as compared with controls. Prominent immune response was observed to a peptide epitope derived from prostaglandin D2 receptor (DP1), as well as peptides homologous to B cell lymphoma 6 protein. Further validation confirmed that these can act as true antigenic targets in discriminating NT1 diseased along with a novel epitope of hemagglutinin of H1N1 to delineate exposure to H1N1.
Interpretation
We propose that DP1 is a novel molecular target of autoimmune response and presents a potential diagnostic biomarker for NT1. DP1 is involved in the regulation of non-rapid eye movement (NREM) sleep and thus alterations in its functions could contribute to the disturbed sleep regulation in NT1 that warrants further studies. Together our results also show that MVA is a helpful method for finding novel peptide antigens to classify human autoimmune diseases, possibly facilitating the design of better therapies.
Keywords: Narcolepsy type 1, H1N1, Pandemrix, Antibody, Prostaglandin, DP1
Highlights
-
•
Using large-scale antibody profiling, differences between narcolepsy type 1 (NT1) and healthy controls became evident.
-
•
Prostaglandin D2 receptor DP1 was delineated as a novel autoantigenic target in NT1.
-
•
Utility of a 3-biomarker ELISPOT assay to assess exposure to H1N1 influenza virus and susceptibility to NT1 was demonstrated.
This study reports on the diagnostic value of novel clinical biomarkers for detecting spontaneous and Pandemrix vaccine-associated NT1 and for stratification of A/H1N1 flu-specific response. Large-scale antibody profiling approach revealed the broad scope and high heterogeneity of NT1-associated immune response. We found a strong autoimmune-like response to prostaglandin D2 receptor DP1 suggesting a firm link between DP1/PDG2 and histamine pathways and the molecular basis of NT1. Together our results warrant further investigation of autoimmune ("self-") response in particular to PDG2 biosynthesis pathway as antibodies to the latter may modify the function of pharmaceutical compounds aimed at treating NT1.
1. Introduction
Narcolepsy type 1 (NT1) is a chronic neurological disease characterized by irresistible daytime sleepiness, disturbed nocturnal sleep, and cataplexy associated with the inadequate function of the hypothalamus (Peyron et al., 2000; Thannickal et al., 2000; Partinen et al., 2014). The major neuropathological features of NT1 are loss of orexinergic neurons and an increased gliosis in the posterior hypothalamic nuclei (Partinen et al., 2014). Increased levels of pro-inflammatory cytokines have been associated with (spontaneously occurring) idiopathic (sNT1) and Pandemrix vaccine-induced narcolepsy (Pdmx-NT1) close to disease onset (Lecendreux et al., 2015). Pandemrix (Pdmx) is an influenza vaccine used during the H1N1 2009 swine influenza A(H1N1) pandemic and was distributed to over 30 million people in EU/EEA countries during the A(H1N1) outbreak. As of January 2015, >1300 cases of vaccine-associated NT1 had been reported to the European Medicines Agency. Epidemiologic and clinical studies conducted in different countries including Finland, Sweden, Ireland, England, Norway, and France have confirmed the association of NT1 in children and adolescents with the AS03-adjuvanted Pdmx (Partinen et al., 2014; Sarkanen et al., 2017). Subsequently, wild-type influenza A(H1N1) infections in China were associated with narcolepsy (Han et al., 2013, 2011). Along with the pandemic A(H1N1) infection, seasonality and post-infectious priming by upper respiratory tract viruses and streptococci have been suggested as triggers of autoimmune response that leads to NT1 in genetically susceptible individuals (Aran et al., 2009; Longstreth Jr et al., 2009).
Genome-wide association studies have revealed a strong association of narcolepsy with the T-cell receptor alpha locus (Hallmayer et al., 2009) and especially with Major Histocompatibility Complex (MHC) class II DQB1*06:02 alleles (Bonvalet et al., 2017; Tafti et al., 2014). DQB1*06:02 is present in approximately 30% of Finnish and Swedish populations (Bomfim et al., 2017). In Finland, all patients with Pdmx-NT1 have been positive for DQB1*06:02 (Partinen et al., 2014). The latter immune haplotype is also strongly associated with the Pdmx-NT1 in Sweden (Bomfim et al., 2017). In another series of 522 patients with narcolepsy and cataplexy from different countries, only 9 patients (1.7%) with low levels of orexin (OX) in cerebrospinal fluid (CSF) were DQB1*06:02 negative (Han et al., 2014). It was also suggested that cross-reactive epitopes to Pdmx vaccine antigens may exist in NT1 diseased as a significant proportion of HLA-DQB1*0602–positive Finns diagnosed with NT1 and with a history of H1N1 vaccination were immunoreactive to OX receptors (Ahmed et al., 2015). However, it still is unclear whether OX-positive neurons and/or their neighboring cells express OX receptors that could be targets for the immune response in NT1 (Valko et al., 2013; Vassalli et al., 2015). The antibody levels to viral nucleoprotein (NP), a Pdmx vaccine antigen, were increased in NT1-diseased carrying the HLA DQB1*06:02 allele (Vaarala et al., 2014), whereas the role of this and other circulating (including intrathecal) autoantibodies in NT1 pathogenesis is not fully understood (see list of previously identified antigens in Table S1). Although NT1-related autoantibodies are found in some patients, the clinical response to intravascular immunoglobulin (IVIG) has been hard to predict (Knudsen et al., 2012). Likewise, use of the drug rituximab might have only short-lasting beneficial effects in NT1 (Sarkanen et al., 2016).
Recent advances in proteomics (immunomics) have made it possible to study the adaptive immune response in various diseases in great detail and at a high resolution (lately reviewed in: (Ayoglu et al., 2016, Wu et al., 2016). We and others have suggested a strategy of high-throughput sequencing-assisted epitope mapping directly on serum for biomarker discovery and disease detection based on the idea that self- and environmental (exposome) antigens are reflected in the immune response profiles (immunoprofiles) (Anastasina et al., 2017; Christiansen et al., 2015; Ionov, 2010; Xu et al., 2015). Hence, the profiling of antibody response repertoire with high-density random peptide/polypeptide display methods could be a novel mean to characterize and classify human diseases in an unbiased manner according to the molecular/cellular targets relevant for the disease.
In the present study, we have used the mimotope-variation analysis (MVA) method to immunoprofile autoantibody repertoires in patients afflicted by NT1 and in controls. We had access to the clinical cohorts composed of 16 NT1 (sNT1 (n = 6) and Pdmx-NT1 (n = 10)) cases, where all NT1-diseased subjects carried the HLA DQB1*06:02 allele, and apart from 2 sNT1 patients, all had been vaccinated with Pdmx. For reference, we used three well-defined control groups: 16 Pandemrix-vaccinated healthy controls (Pdmx-HC), 16 H1N1-infected Finnish subjects (H1N1-HC), and 16 healthy Estonian donors (HC – healthy controls) (Table 1). Our data revealed complex patterns of immune response in all patient groups including novel epitope sequences present in sera of Pdmx-NT1 and H1N1-HC. One such peptide epitope was identified as belonging to the prostaglandin D2 receptor (DP1) that together with its ligand prostaglandin D2 (PGD2) is involved in sleep regulation in humans and experimental animal models (see ref. in Urade and Hayaishi (2011)).
Table 1.
Description of samples studied.
| Characteristics | Narcolepsy (NT1) patients |
Healthy controls (HC) |
|||
|---|---|---|---|---|---|
| Pdmx-NT1 | sNT1 | Pdmx-HC | H1N1-HC | Other HC | |
| Group size (n) | 10 | 6 | 16 | 16 | 16 |
| Gender (female/male) | 5/5 | 5/1 | 12/2a | 0/16 | 10/6 |
| Pandemrix vaccination | 11/2009–1/2010 | 11/2009-1/2010b | 11/2009-1/2010 | – | – |
| Sample collection | 2011 | 2011 | 2011 | 2011 | 2009 |
| Median age at onset (y) | 13 | 18 | – | – | – |
| Median age at sampling (y) | 14 | 22 | NA | 21 | 34.5 |
| Unambiguous cataplexy | 10/10 (100%) | 6/6 (100%) | – | – | – |
| MSLT mean SL (range) | 2.0 (0.4–4.3) | 2.6 (0–7.5) | NA | NA | NA |
| SOREMPS mean (range) | 3.7 (2–5) | 2.7 (2–4) | NA | NA | NA |
| HLA DQB1*0602 (%) | 10/10 (100%) | 6/6 (100%) | NA | NA | NA |
| CSF-orexin < 150 pg/mL (lower 1/3 limit in Finland) | 7/7 (100%) | 5/5 (100%) | NA | NA | NA |
HC- healthy control, H1N1-HC – H1N1 infected, Pdmx-HC– Pandemrix-vaccinated, NT1- narcolepsy type 1 (including 10 Pdmx-induced NT1 samples (Pdmx-NT1) and 6 sporadic NT1 (sNT1) samples), NA – not available, SL – sleep latency, MSLT – Multiple sleep latency test, SOREMPS – Sleep onset REM periods as defined by the American Academy of Sleep Medicine.
Gender of two Pdmx-HC is unknown.
Four out of 6 sNT1 patients were vaccinated after they had been diagnosed with NT1.
2. Materials and Methods
2.1. Vaccines
Pandemrix vaccine is derived from X-179A, a reassortant of hemagglutinin (HA), neuraminidase (NA) and polymerase acidic protein (PA) of A/California/07/2009 and X-157 H3N2 in a PR8 backbone (Jacob et al., 2015; Nicolson et al., 2012; Robertson et al., 2011). The vaccine composition can be found summarized by European Medicines Agency and GlaxoSmithKline plc (European Medicines Agency, 2009).
2.2. Study Population
The present study comprises a total of 64 individuals (Table 1). Altogether, 16 serum samples of H1N1-infected military servicemen (H1N1-HC), 16 serum samples of age/sex-matched Pandemrix-vaccinated healthy controls (Pdmx-HC) were kindly provided by National Institute of Health and Welfare, Finland. 16 serum samples were collected from patients with H1N1-induced (Pdmx-NT1) and sporadic narcolepsy (sNT1). Four out of 6 sNT1 patients were vaccinated with Pdmx after they had been diagnosed with NT1. Narcolepsy patients were diagnosed at the Finnish Narcolepsy Research Center (Helsinki Sleep Clinic, Vitalmed Research Center) by experienced neurologists with a special competence in sleep medicine. All narcolepsy patients had NT1 as defined by the American Academy of Sleep Medicine international criteria of sleep disorders version 3. All excessively sleepy patients with NT1 had unambiguous cataplexy and an abnormal MSLT (sleep latency <8 min and at least 2 sleep onset REM periods; Multiple Sleep Latency Test) recording after a polysomnography. Twelve of the 16 patients had their CSF-orexin levels measured using the standardized Phoenix RIA method with Stanford reference. All except one had CSF-orexin levels <110 pg/mL (one woman with clearly abnormal MSLT and unambiguous cataplexy had CSF-orexin level of 127 pg/mL).
Control serum specimen for the study included 16 serum samples from Blood Centre, North Estonian Regional Hospital, Estonia, collected in 2009 prior to the swine influenza pandemic (other HC). Sera were stored at −135 °C until use.
2.3. Ethical Permissions
The patients have participated in the NARPANord narcolepsy study (Academy of Finland, grant nr. 260603), and they have given a written informed consent. The serum samples of H1N1-infected military servicemen and serum samples of the Pdmx-vaccinated healthy controls were provided by the National Institute of Health and Welfare, Finland. The ethical permissions were approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa, Finland.
2.4. Mimotope-Variation Analysis
For qualitative and quantitative characterization of humoral immune response from sera samples, we used an in-house developed mimotope-variation analysis (MVA) method. Fig. 1A provides an overview of the process. In brief, a random 12-mer peptide phage library (Ph.D.-12, NEB, UK) was used according to the manufacturer's protocol. 2 μL of serum sample was incubated with 2.5 μL library (~5 × 1011) and immunoglubulin G (IgG) fraction was recovered using protein G-coated magnetic beads (Thermo Fisher Scientific). The unbound phage particles were removed by extensive washes with TBS-T (TBS + 0.1% [v/v] Tween-20). Selectively captured phage DNA was analyzed by using next generation (Illumina) sequencing with barcoding primers (Islam et al., 2014). For that, DNA was extracted by using standard NaI/EtOH precipitation method and enriched by PCR amplification using primers enriched with adapters for the sequencing reaction that flank the variable region at the end of pIII in M13KE vector (Fw: 5′-AATGATACGGCGACCACCGAGATCTACACTGATCTAGTGGTACCTTTCTATTCTCA*C*T*C*T-3′ Rv: 5′-CAAGCAGAAGACGGCATACGAGATNNNN(NN)CCCTCATAGTTAGCGTAACG-3′). PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN), and the concentration of DNA with Illumina adapters was estimated by Qubit Fluorimeter (Invitrogen) according to the manufacturer's protocol. Sequencing was performed using Illumina HiSeq and 50-bp single end reads. Samples were analyzed at least in duplicates to ensure reproducibility. To evaluate the data reproducibility, we compared peptide abundance in two replicates using Pearson's correlation coefficient test (r value higher than 0.90, p < .0001).
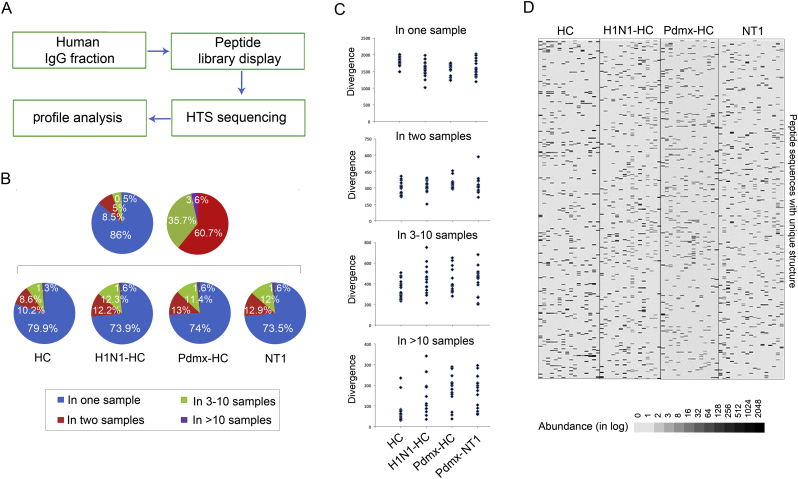
Fig. 1.
Humoral immune response studied using the mimotope-variation analysis (MVA) method. A. Schematic drawing of the workflow in MVA. MVA is a high-throughput random peptide phage display analysis. A random peptide display library (PhD12) was used which contained 10^9 different 12-mer peptide sequences introduced to the N-terminus of the phage major coat protein pIII (NEB). For MVA, sample-specific IgG proteins (antibodies, Human IgG fraction) present in human sera of interest are allowed to interact with the phage-displayed peptides and the IgG-phage complexes were captured to protein G magnetic beads, while the unbound phages were washed away (Peptide library display). Captured phages were lysed and DNA amplified with primer sequences containing a tag with a unique barcode sequence and the final amplicons were pooled for NGS analysis (HTS sequencing). The primer set homologous to the M13KE vector sequences that flank the random peptide coding sequence was used to amplify a 50-bp fragment. Data analysis to classify peptides that were specific to Pmdx-infected, -vaccinated and NT1-diseased individuals was carried out by comparing the profiles of peptides (mimotopes) from diseased to those from non-diseased (Peptide profile analysis). On average, MVA generated 1.8 million peptide sequences with unique structure (divergence) totaling 2.8 million peptide sequences in abundance (total abundance; number of reads) per sample. Altogether, a peptide data set with >16 million sequences (Totpep) with unique structure was generated.
B. Analysis of peptides revealed highly divergent patterns (immunoprofiles) across study cohorts. The fraction of top 2500 peptides with unique structure and highest values of abundance - reflecting the peaking immune reactivity of each sample - was analyzed for variance. Top2500 peptide dataset contained altogether 160,000 sequences out of which 121,142 were unique. Pie charts display the sequence distribution of unique peptides across all samples analyzed. The left pie (blue) displays the proportion of shared vs. unique peptides: ~86% were unique to one individual whereas ~14% of the peptide sequences were shared between samples, out of these ~8.5% were common to 2 samples, 5% to 3–10 samples and 0.5% were detected in >10 samples. The right pie (red) displays the distribution of shared 16,844 peptide sequences out of which ~60.7% were common to 2 samples, 35.7% to 3–10 samples and 3.6% were seen in >10 samples. The four pie charts (below) exemplify the peptide profile structures in different clinical cohorts. The size of each pie piece is proportional to the number of unique peptides common to one or more samples of a clinical cohort. Blue - represents unique peptides, red - the most shared.
C. Individual variation in peptide divergence is characteristic to all immunoprofiles. Top 2500 peptides were analyzed to assess the range of individual peptide variation across study cohorts. Blue dots mark peptide divergence in a single sample. As indicated, between one to two thousand peptides were individual-specific, whilst the most common peptides (shared by >10 individuals) ranged in divergence from tens to 350 across samples. Range of unique peptide variations was similar across all study samples.
D. Heat map image of a random fragment of MVA profile encompassing 400 peptides across study samples. Peptide profiles were individual-specific with a highly varying abundance. Each column represents the peptide profile of a single individual, and each line represents a peptide with a unique primary structure. Abundance is presented as counts in logarithmic scale (in log); black colour depicts peptides captured at higher abundance, and white those at lower abundance. Shown are peptide profiles that were common to 3–10 individuals. Abbreviations: Abundance – peptide frequency; Divergence – all unique peptides; HC- healthy control; H1N1-HC – H1N1 infected; Pdmx-HC- Pandemrix-vaccinated; NT1- narcolepsy type 1 (including 10 Pandemrix-induced NT1 samples).
2.5. Sequence Data Analysis
Every read that was considered valid by the Illumina HiSeq control software was prepared for further analysis by trimming. In brief, sequence reads of 50 bp were accompanied with a 4-bp tag to assign each read uniquely to one out of 48 multiplexed samples. After demultiplexing there were millions of sequence reads for each sample. Each read consisted of a 36-bp random insert region and a constant region. We discarded the reads with mismatches in flanking 4 bases of the constant region. Sequences from wild-type phages with no random insert were excluded. Next, we translated all the random insert regions in reads into peptide sequences of length 12aa. All non-translatable sequences were discarded. To reduce the effect of amplification and sequencing errors, only those peptide sequences were kept that had at least two copies sequenced per sample. In order to compensate for the different numbers of reads per sample normalization of read counts was performed. All samples were trimmed to 3 million reads (RPM units). The resulting data was represented as a cross-table where each row corresponded to a different 12mer peptide, each column corresponded to a different sample, and each cell showed the read count of the peptide in the respective sample measured in RPM-units. According to the manufacturer (NEB), naïve library contained up to 109 different sequences. For reasoned cost purposes, the estimated outcome of sequence data represented 0.1% of the initial library input containing up to 2.8 × 106 different peptide sequences per sample. Complete analysis of sequence diversities obtained by MVA remains out of the scope of the current study.
2.6. Clustering Workflow
The main assumption was that every obtained peptide sequence mimics the target of an antibody. The sequence reads of one sample often included many copies of the same peptide sequence. The read counts of a peptide could range from 1 to thousands. To reveal recognition patterns (epitope motifs) which were enriched in the cases compared to controls, we used SPEXS2 software (https://github.com/egonelbre/spexs2; (Vilo, 2002, Brazma et al., 1998)). For clustering the peptides with motifs and generating mimotope regular expression and sequence logos, the “motifTree” tool was used (Kruup, 2013). The Multiple EM for Motif Elicitation (MEME-MAST) algorithm (Bailey and Elkan, 1994; Bailey and Gribskov, 1998) was used to align peptides to proteins. For B cell epitope mapping IEDB 3.0 database was used (Vita et al., 2015).
2.7. Statistical Analysis
All statistical analyses (ANOVA, t-Test, correlation analyses, Chi-square test) were done using MedCalc software (MedCalc Statistical Software version 17.0.4 (MedCalc Software bvba, Ostend, Belgium; https://www.medcalc.org; 2017)). For visualization of peptide abundance across samples, peptide frequency values were converted to heatmap images (Tagged Image File) with Excel Visual Basic for Applications (VBA) scripts. For visualization of selected peptide set alignment profile on proteins of interest Excel VBA script was used. The protein sequence was scanned with every peptide and at every position where the peptide aligned with it in at least four perfectly matching positions, one was added with its frequency. For random reference profile, amino acid sequence of each peptide was randomized and scanned using the same rules over the target sequence.
2.8. Influenza Virus Serology
Levels of influenza-specific IgG antibodies were determined by the enzyme-linked immunosorbent Vir-ELISA anti-H1N1/H3N2 IgG assay (Influenza virus type A IgG ELISA test system, Euroimmun), carried out in accordance with the manufacturer's specifications. Absorbance was measured at 450 nm with SpectraMax Paradigm.
2.9. Peptide ELISPOT
For peptide ELISPOT the following peptides were designed:
peptide #1 - RVLAPALDSWGTGGGDYKDDD{LYS(BIOTIN)} (Genescript)
peptide #2 - LPKFSAPSASGPGGGDYKDDD{LYS(BIOTIN)} (Genescript)
peptide #3 - ESTRYQLWLPHQGGGDYKDDD{LYS(BIOTIN)} (Genescript)
control peptide - AVLAAALASWGTGGGDYKDDD{LYS(BIOTIN)} (Genescript)
In brief, 110 pg biotin-conjugated peptides were printed on nitrocelluose coated slides (10485323, Whatman) by SpotBot® 4 (Arrayit). For primary antibody human precleared serum (1:100) was used, for secondary antibody rabbit anti-human IgG (H&L) (HRP) (Abcam) was used. All incubations were done for 1 h at room temperature. Results were scanned using Ettan DigeImager (GE Healthcare Life Sciences) and images calculated using ImageQuant software version 8.1 (GE Healthcare Life Sciences).
2.10. Cancer Cells, Human Mesenchymal Stem Cells (hMSC) and Post-Mortem Tissues
Immortalized glioblastoma multiforme cells (human glioma cells - hGC) (kind gift of Prof. Aavo-Valdur Mikelsaar, Estonia), human neuroblastoma cell line Kelly (ATCC) and human mesenchymal stem cells (hMSC) (isolated from human subcutaneous adipose tissue as described (Jaager and Neuman, 2011)) were grown in Dulbecco's modified Eagle's medium (DMEM (PAA)) containing 10% fetal bovine serum (PAA), 1 mg mL−1 penicillin (PAA) and 0.1 mg mL−1 streptomycin (PAA). All cells were cultured at 37 °C in 5% CO2. The identity of hMSC was confirmed by using cell morphology and flow cytometry methods for analysis of cell surface markers: CD73+/CD90+/CD105+/CD45−/CD34− (Kauts et al., 2013). For treatments, hMSCs were grown with media containing IL-1β (1 ng/mL), IFNγ (2 ng/mL) for 8 h, or PGD2 (10 μM) for 1 h.
Human post-mortem tissues were procured from the North-Estonian Regional Hospital, Tallinn, Estonia. All experiments with human tissues were done with the approval of the local ethical committee (license no. 2234, date of issue 09.12.2010).
2.11. Immunofluorescence and Western Blot Analysis
For immunofluorescence analysis, cells grown on glass inserts were fixed using 4% PFA (Scharlau) for 15 min and blocking of the unspecific reactivity was done with 5% BSA. The antibodies used included: anti-DP1 (Abnova; 1:500), precleared human sera (1:400), and the secondary Alexa Fluor 488 and 647 (Invitrogen, 1:2000) antibodies. For epitope blocking peptide #1 was used in final concentration 6.6 μg/mL. Hoechst 33342 (Invitrogen) was used to detect cell nuclei. Imaging was done using Nikon Eclipse 80i microscope.
Sequences (RVLAPALDSWGT and DYKDDDDK (flag)) were inserted at the N-terminus of the pIII of the M13KE phage by in vitro mutagenesis PCR using primers s1 5′GCTGGATAGTTGGGGAACCGGTGGAGGTTCGGCCGAAAC3′, as1 5′GCCGGAGCTAGTACACGAGAGTGGGAGTAAAACGGTACC3′,
s2 5′GCTGGATAGTTGGGGAACC3´, as2 5`GCCGGAGCTAGTACACG3′; s3 5′GATGATGATAAAGGTGGAGGTTCGGCCGAAAC3′, as3 5′ATCTTTATAATCAGAGTGGGAGTAAAACGGTACC3′; s4 5′GATGATGATAAAGGTGG3′, as4 5′ATCTTTATAATCAGAGTGG3′. PCR reactions were carried out with phusion Hot Start II High-Fidelity DNA Polymerase (ThermoScientific). Constructs were verified by sequencing. For Western blot analysis, 30 μg of protein lysate or 1 × 1013 phage particles were resolved on 10% SDS–polyacrylamide gels and transferred onto PVDF membranes (Amersham) for 1.5 h using BioRad wet blotter in standard Towbin buffer. The membrane was blocked with 5% nonfat milk (AppliChem), incubated overnight with the following primary antibodies: anti-DP1 (St. John Laboratory, 1:1000), anti-GAPDH (Sigma, 1:10,000), precleared human sera (1:500). The epitope blocking peptide #1 was used in final concentration 6.6 μg/mL. The membrane was incubated for 1 h at room temperature with the secondary anti-mouse, anti-rabbit, or anti-human IgG antibodies (Abcam; dilution 1:10,000). The ECLfemto kit (Amersham) was used for detection of immunoblotted target proteins.
2.12. RNA Extraction, RT-PCR and qRT-PCR
Total RNA from human brain parts was extracted using RNAWiz (Ambion) as recommended by the manufacturer. Total RNA from cells was isolated using TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions. One microgram of RNA was reverse transcribed into cDNA using SuperScript III first strand cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. The resulting cDNAs were used as templates for subsequent RT-PCR reactions. RT-PCR was carried out using FIREPol® DNA polymerase (Solis Biodyne), 40 amplification cycles and an annealing temperature of 58 °C. Amplification of the housekeeping gene GAPDH was performed for 25 cycles using FIREPol® DNA polymerase (Solis Biodyne) and used as an internal control. Used primer sequences: PTGDR sense 5′ATGAAGTCGCCGTTCTACC3′, PTGDR antisense 5′CATGAAGAAGGCGAAGGCTTG3′, GAPDH sense 5′GAAGGTGAAGGTCGGAGT3′, GAPDH antisense 5′GCATGGACTGTGGTCATGAG3′. IL-1β sense 5′GGGCCTCAAGGAAAAGAATC3′: IL-1β antisense 5′TTCTGCTTGAGAGGTGCTGA3′, IFNγ sense 5′CTGTTACTGCCAGGACCCAT3′, IFNγ antisense 5′TTTCTGTCACTCTCCTCTTTCCA3′.
3. Results
3.1. Autoimmune Response Profiles Across Cohorts are Highly Heterogeneous
We performed MVA by selecting peptide antigens from random phage library (PhD12, NEB) with 10^9 different 12-mer peptide sequences based on their high avidity of interaction to antibodies in sera (Fig. 1A). A total dataset of 16 million peptides with unique sequences was generated. The data structure analysis of Top2500 peptide dataset (the most abundant peptides across individual samples) revealed that although these peptides were largely individual-specific (Fig. 1B and D), the study cohorts shared a fraction of common characteristics across Top2500 features (Fig. 1B). The remarkable heterogeneity of antigenic reactivity between individuals has also been noted previously (Zandian et al., 2017). However, the distribution of peptides according to the frequency was found to be similar in the different clinical subsets (Fig. 1C).
3.2. H1N1-specific Immunoprofiles are Largely Shared Between Pdmx-Vaccinated and Subjects Infected With H1N1
To evaluate the extent to which the presence of H1N1-specific peptides was restricted to specific clinical subsets, we assayed responses to H1N1 infection and Pdmx-vaccination using type A influenza ELISA (Quantum) diagnostic tests. High-titer responses to influenza A virus major antigens (including H1N1) were evident for both Pdmx-vaccinated and H1N1 naturally infected individuals (Fig. 2A). The humoral response to seasonal flu (A/H1N1 and A/H3N2) was relatively weaker in NT1-diseased as compared with Pdmx-HC individuals as determined by using a commercial ELISA test (p < .001). This was in slight contrast to earlier findings reporting that Pdmx-NT1 patients had higher median levels of anti-H1N1 antibodies than controls (Lind et al., 2014), and may reflect the characteristics of the samples collected (see Table 1, Materials and methods). Next, we assessed the reactivity of the sera to protein fragments representing the four major antigens of H1N1 virus proteome (strain A/California/7/2009). MVA data analyses of Top2500 peptide data set revealed 4 antigenic regions for hemagglutinin (H1N1/HA, C4RUW8), 5 for neuraminidase (H1N1/NA, C3W6G3), 3 for nucleoprotein (H1N1/NP, B4URE0), and 6 for polymerase acidic protein (H1N1/PA, I6THC5), some of which corresponded to known immunogenic epitopes from IEDB (http://www.iedb.org/; Fig. 2B). Statistically distinct coverage profiles with different peaks on H1N1 HA, NA, NP, and PA antigens were obtained from analysis of Top2500 peptide data sets of H1N1-HC, Pdmx-HC and NT1 samples (Fig. 2C). Data showed that the most commonly shared epitopes raised by the anti-Pdmx/anti-H1N1 immune response were found in the C-terminal region of H1N1/HA (C4RUW8) locating between amino acids 521 to 531 (Fig. 2B–C), directly before a proven T cell-antigenic region in HA between amino acids 527–541 of A/California/04/2009 (H1N1, (Schanen et al., 2011)). About 700 peptides from the total peptide dataset clustering to motif with sequence consensus E[ST].R.[QM] were highly abundant in H1N1-HC, and relatively infrequent in Pdmx-HC and NT1 samples as compared with HCs (Fig. 2D).
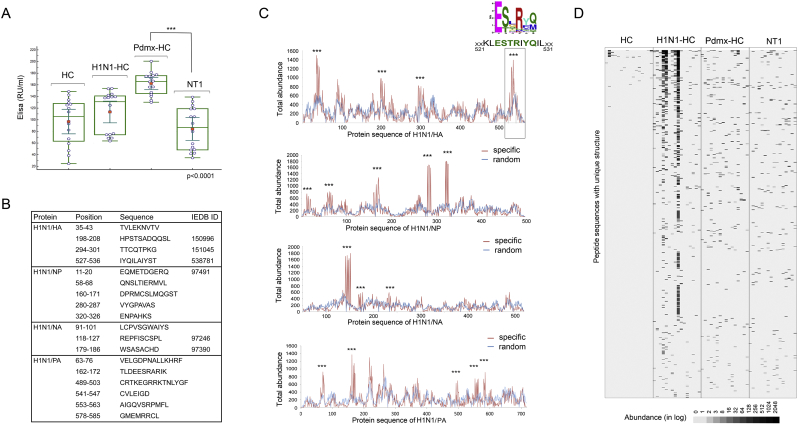
Fig. 2.
A novel common epitope of HA antigen of A/H1N1 in seasonal infection carriers and Pdmx-vaccinated individuals, encompassing a proven T cell- antigenic region.
A. The humoral response to seasonal flu (A/H1N1 and A/H3N2) was relatively weaker in NT1-diseased as compared with Pdmx-HC individuals as determined by using a commercial ELISA test (p < .001). The presence of IgG antibodies against HA and H3N2 was assessed in sera samples of HC, H1N1-HC, Pdmx-HC and Pdmx-NT1 by ELISA (Influenza virus type A IgG ELISA, Euroimmun). The HC samples were collected prior to A/H1N1 outburst in Estonia, before the fall 2009. Blue circles mark individuals of the study cohorts; red dots mark the mean values; lines depict median values; inner whiskers mark confidence interval for the mean; boxes mark upper and lower quartiles; outer whiskers mark the maximum and minimum values (excluding the outliers). P-values were calculated by ANOVA and are marked with asterisks. The cut-off value for ELISA was 16 RU/mL. Labels at the top of box plots demark the clinical origin of the sample.
B. MVA predicted H1N1 epitopes partially overlapped with previously described H1N1 (A/California/08/2009(H1N1)) B cell specific epitopes from IEDB (http://www.iedb.org/). Top2500 peptide dataset containing 121,142 unique sequences was used to delineate the predominant epitopes of H1N1/HA (GI: 238,623,304), H1N1/NA (NA, GI:758899360), H1N1/NP (NP, GI:229891180) and H1N1/PA proteins in study samples. 9657 peptides from the studied dataset satisfied the selection criteria that these were not present in the HC samples. Specific alignment profiles for each of the A/H1N1 protein antigens were calculated with the criterion that the abundance of a peptide was to be 2-fold higher over random.
C. MVA immunoprofiles predicted a novel epitope in the C-terminal region of HA encompassing amino acids 521–531 and with the sequence ESxRxQ that was common to both seasonal infection carriers and Pdmx-vaccinated individuals. The graphs show antigen-specific profiles of overall peptide abundance where the number of peptides were counted for each amino acid position for the following proteins: hemagglutinin (H1N1/HA, C4RUW8), neuraminidase (H1N1/NA, C3W6G3), nucleoprotein (H1N1/NP, B4URE0) and polymerase acidic protein (H1N1/PA_I6THC5). Amino acid sequence of the proteins is depicted on the x-axis. Marked with asterisks are regions where set calculation criteria were satisfied. Detailed analysis of immunoprofiles of H1N1 antigens revealed a novel immunogenic region of HA encompassing amino acids 521–531 that corresponds to the earlier experimentally determined A/Puerto Rico/8/1934(H1N1) HA520–530 CTL epitope (Gianfrani et al., 2000) and is partially overlapping with broadly reactive CD4+ T cell epitope: HA527–541 of A/California/04/2009(H1N1) (Schanen et al., 2011). Peptides aligning to 521–531 of HA cluster to a minimal consensus sequence E[ST].R.[QM] by sequence homology alignment.
D. Heat map image of immunoprofiles of peptides with consensus E[ST].R.[QM] across study samples. The total peptide data set was examined for the peptides with unique structure clustering to E[ST].R.[QM] motif. About 700 peptides with enriched abundance in H1N1-HC, Pdmx-HC or Pdmx-NT1 samples were found to cluster to the motif. The data of 700 peptides is presented on the heat map image. Each line represents peptides with unique sequence structure. The colour intensity of each cell corresponds to the peptide abundance (presented in log value). Black represents peptides captured at higher abundance whereas white represents peptides captured at lower abundance. Each column represents a peptide profile from a single sample. Labels at the top of the panels indicate the clinical origin of the sample. Abbreviations: Random alignment – amino acid sequences of peptides under analysis were randomized and aligned to respective protein coding sequence; Total abundance - the number of peptides counted for defined amino acid positions; HC– healthy control; H1N1-HC – H1N1 infected; Pdmx-HC- Pandemrix-vaccinated; NT1- narcolepsy type 1 (including 10 Pandemrix-induced NT1 samples).
3.3. Examination of Identified NT1-specific Autoantigens in MVA Dataset
Next we determined peptides that were different between the clinical study groups to examine whether they were consistent with the prior knowledge of Pdmx-NT1-specific immunogenic epitopes. For the study, we used an exhaustive sequence pattern search (SPEXS - https://github.com/egonelbre/spexs2; (Vilo, 2002, Brazma et al., 1998)) gene ontology analysis, combined with the interrogation of the presence of known autoantigens previously identified in Pdmx-NT1 disease (Table S1(Ahmed et al., 2015, Bergman et al., 2014, Cvetkovic-Lopes et al., 2010, De La Herran-Arita et al., 2013, Haggmark-Manberg et al., 2016, Katzav et al., 2013, Zandian et al., 2017)). Thus, we were able to confirm statistically significant patterns of epitope recognition in the samples. Particularly, we identified epitopes resembling those in the N-termini of OX (4/16) and OX1R/2R (2/16; 4/16), in mitogen-activated protein kinase 7 (MAP3K7) (amino acids 318–328; 3/16) and in 5′-nucleotidase cytosolic IA (NT5C1A) (amino acids 35–48; 2/16), as well as in B-cell lymphoma 6 protein (BCL6), encompassing amino acids 279–288 in 6 out of 16 sera samples of NT1 diseased (Fig. 3, Table S1). According to MVA data, none of the previously identified antigens was prominently detected across NT1 diseased and were also common also to HC if less stringent statistical power criteria were used (Fig. 3). In contrast, we found no evidence of stratifying peptides with consensus sequences mimicking tribbles pseudokinase 2 (TRIB2), neuropeptide glutamic acid- isoleucine/α-melanocyte-stimulating hormone (NEI/aMSH), or others that were reported by earlier studies (Table S1). These data allowed concluding that apart from the BCL6 related subset, peptides corresponding to previously identified autoantigens had relatively little discriminative power, suggesting also that these antigens were either rare or recognized promiscuously in patient groups with a clinical and ethnical heterogeneous background.
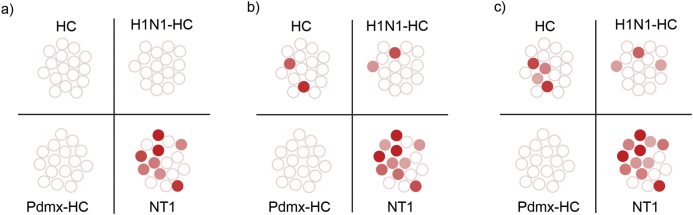
Fig. 3.
Heterogeneity of the immune response to delineated antigens is apparent at the individual level. The total peptide data set was examined for peptides with homology to different antigens (see Table S1). Homology alignment analysis resulted in delineating six minimal consensus motifs with homologies to epitopes defined by previous studies. Namely, LPxTNxS (HCRT, O43612), RDxxYP (HCRTR1, O43613), SxLNxTxN (HCRTR2, O43614), KxxxPSAS (BCL6, P41182), STNxS (MAP3K7, O43318), LAxSxKP (NT5C1A, Q9BXI3), (Table S1). Datasets from different study cohorts were analyzed for the presence of peptides containing the 6 motifs and ROC analyze was performed at threshold values: a) specificity 100% (sensitivity 50%), b) specificity 90%, (sensitivity 63%), c) specificity 85% (sensitivity 69%). Note that the ensemble snapshots from MVA data did not distinguish between the possible scenarios of each individual antigen motif in single individuals. Individual samples are colour coded. White circles indicate that peptides with abundance values clustering to one or more aforementioned motifs were below threshold. Colored circles indicate peptides with abundance values clustering to one or more aforementioned motifs were above threshold. None of the motifs displayed a statistically significant association with many other known antigens (see the list of antigens in Table S1). This negative result may be due to a limited statistical power of delineated antigens, but also due to the heterogeneity of autoantibody repertoires in different individuals. Abbreviations: HC- healthy control; H1N1-HC – H1N1 infected; Pdmx-HC- Pandemrix-vaccinated; NT1- narcolepsy type 1 (including 10 Pandemrix-induced NT1 samples).
3.4. A Defined Set of Peptides Derived From DP1 Acts as Antigenic Epitopes in NT1
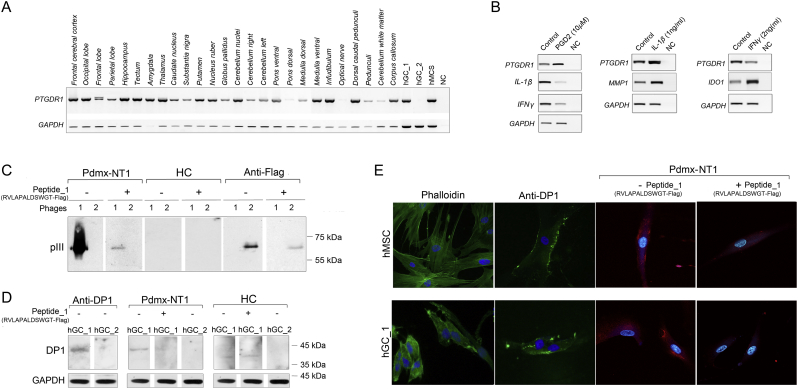
In analyses of the peptides that were unique among the disease groups, we observed that the Top2500 dataset contained >1300 peptides with a high enrichment in NT1 (Fig. S1A–B). The most abundant peptide having the sequence RVLAPALDSWGT showed a high sequence homology within the second extracellular loop region in the human prostaglandin D2 receptor DP1 (Q13258). This region in DP1 is predicted to function in ligand recognition (Avlani et al., 2007; Nagata et al., 2017) and is not conserved in mouse and rat (Fig. 4A). Extraction of all peptides from the total dataset having the highest homology to RVLAPALDSWGT and to DP1 revealed a set of 4428 unique peptides containing the RxxxPxxD (RPD) consensus sequence that discriminated the NT1 samples from controls (p < .0001, ANOVA, Fig. 4B–C). We then also determined that the 2157 RPD-containing peptides out of 4428 (Fig. 4B) had a high sequence homology to DP1 protein where the bona fide immunodominant epitope with sequence RVLAPALD encompassed amino acids 94 to 101 in DP1 (Fig. 4D). Interestingly, according to the IEBD database (www.iedb.org/), four MHC-I binding epitope regions of DP1 encompassing amino acids 132–140 (ID: 716767), 145–156 (ID: 637966), 195–203 (ID: 727099) and 303–311 (ID: 697995) have been defined. The latter (303−311) encoded another extracellular domain of DP1 that was also defined by us a potential target of B cell response (Fig. 4D).
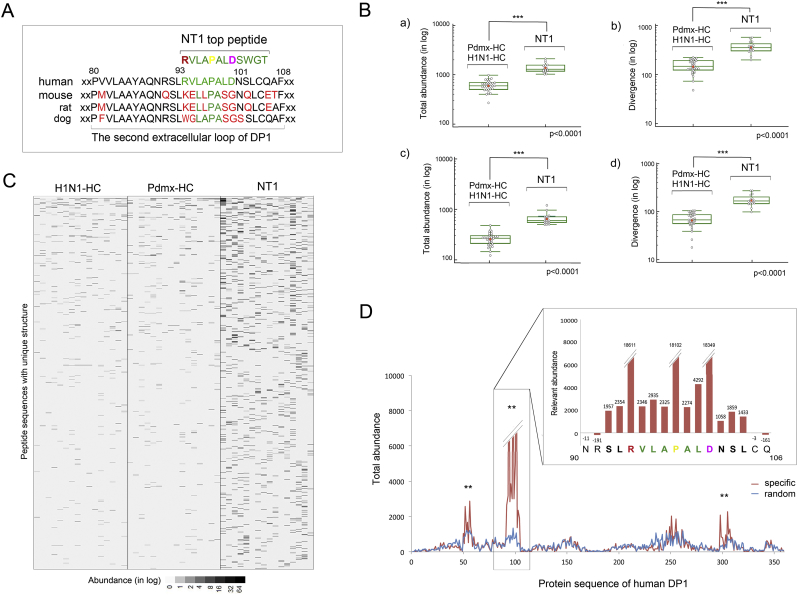
Fig. 4.
Peptides with minimal epitope motif RxxxPxxD (RPD) are enriched in samples of NT1 and align to the extracellular loop of human DP1.
A. Peptide RVLAPALDSWGT found as the most abundant in Pdmx-NT1 was highly homologous to the second extracellular loop region of DP1 protein. More than 1,300 peptides from the selection of the Top2500 peptide dataset were enriched in NT1 samples (Fig. S1A–B) where the peptide RVLAPALDSWGT had the highest values of abundance across the Pdmx-NT1 cohort. BLAST analysis revealed that RVLAPALDSWGT region of DP1 is highly dissimilar in mammals. To identify peptides from Totpep dataset with the highest homology to DP1 and to RVLAPALDSWGT, SPEXS2 software (https://github.com/egonelbre/spexs2.) was used and the top motif that fulfilled both criteria was found to be RxxxPxxD (RPD, a motif defined by three shared amino acids).
B. Peptide sequences containing RPD motif were enriched in immunoprofiles of NT1 samples. Analyzing the Totpep library, 4428 peptides were identified from NT1 (including 10 Pdmx-induced NT1 samples and 6 sNT1 samples) data sets that contained the RPD motif. Box plot depicting that these 4428 peptides (with RVLAPALDSWGT eliminated beforehand as dominant) contained sufficient information to discriminate between HC and NT1 samples in a statistically significant manner a) by their abundance (in log, student t-test p-value < .0001) and b) divergence (in log, student t-test p-value < .0001). SPEXS2 analysis resulted in 2157 peptides that out of 4428 were highly homologous (with at least 4 consecutive amino acid matches) to RVLAPALD of human DP1 and discriminated NT1 samples (including 10 Pdmx-induced NT1 samples and 6 sNT1 samples) in a statistically significant manner c) by their abundance (in log, student t-test p-value < .0001) and d) divergence (in log, student t-test p-value < .0001). Six NT1 samples that were not Pdmx-induced were similarly to Pdmx-NT1 samples discriminated by the set of 4428 peptides from HC. In box plots - blue circles mark the single individuals of study cohorts; red dots mark the mean values; line marks the median values; inner whiskers mark confidence intervals for the mean; boxes mark the upper and lower quartiles; outer whiskers mark the maximum and the minimum values (excluding outliers).
C. Heatmap images depicting the immunoprofiles of the top one thousand NT1-specific peptides out of 4428 across the clinical study-groups. The data are presented as heat map image generated via conditional formatting in MS Excel. Each column represents a peptide profile from a single individual. Each line represents peptides with unique sequence structure. The colour intensity of each cell corresponds to the peptide abundance (counts of sequences in log). Black represents peptides captured at higher abundance whereas white represents peptides captured at lower abundance.
D. Epitope mapping of anti-peptide response to human DP1 in sera of Pdmx-NT1 diseased. SPEXS2 analysis resulted in 2157 peptides carrying RxxPxxxxD motif that were aligned to DP1 protein sequence (Q13258) with a 2-fold higher abundance over random as a chosen criterion. In addition, MEME-MAST algorithm (Bailey and Elkan, 1994) aligned these 2157 peptides to the region encompassing 94-101aa of DP1, E-value = 0.0078. Each bar on the x-axis corresponds to one of the overlapping peptides required to cover the antigen, and the height of the profiles shows the relative abundance. Three potentially immunogenic regions were described with the predominant alignment containing the sequence RVLAPALD and encompassing amino acids 94 to 101. Zoomed in the box is an extract of the immunoprofile of DP1 in positions 90–106. Calculated relative abundance values are marked above each amino acid position. Note that the weaker immunogenic regions of DP1 are expanded toward both N- and C-termini. Abbreviations: Abundance – peptide frequency; Divergence – all unique peptides; HC- healthy control; H1N1-HC – H1N1 infected; Pdmx-HC- Pandemrix-vaccinated; NT1- narcolepsy type 1 (including 10 Pandemrix-induced NT1 samples).
To validate the data, we employed different methods and measured the serologic response to peptides carrying the RPD consensus sequence using sera samples of the study (Fig. 5, Fig. 6). In line with previous reports (Urade and Hayaishi, 2011), we observed a broad expression of DP1 mRNA across different regions of human brain, in human glioma (hGC), and normal mesenchymal stem cells (hMSCs) (Fig. 5A). Treatments of hMSCs with the ligand prostaglandin D2 increased DP1 expression, whilst the pro-inflammatory cytokines interleukin 1β and interferon γ (IL-1β and IFN-γ) either slightly increased or decreased its expression, respectively (Fig. 5B). In contrast, PGD2 strongly reduced IL-1β and IFN-γ expression suggesting that these cells recapitulate the intact PGD2-DP1 signaling pathway by inducing anti-inflammatory responses in the studied cells (Fig. 5B). Performing phage Western blot assay we confirmed that MVA predicted DP1-positive Pdmx-NT1 sera showed IgG reactivity to phages that displayed RVLAPALDSWGT peptides (RVLAPALD-pIII, Fig. 5C). No specific reactivity was detected using DP1-negative sera (Fig. 5C). This peptide target specificity was further confirmed by Western blot analysis using Pdmx-NT1 sera where the interactions between human IgGs and antigen expressing phages were blocked by RVLAPALDSWGT synthetic peptides (Fig. 5C, Fig. S2). Immunoblot analysis using commercial anti-DP1 polyclonal sera and clinical sera of Pdmx-NT1, confirmed the presence of DP1 expression in hGC_1 and not in hGC_2 glial cells and also here specific blocking effects to the seroreactivity in the presence of RVLAPALDSWGT peptides were confirmed (Fig. 5D), but not in case of control peptides or irrelevant sera (Fig. 5C, Fig. S2). Immunocytochemical analyses showed that DP1 was predominantly localized on the cell surface of hMSC and hGC cells, and more importantly, was equally well-detected by immunocytochemistry using commercial anti-DP1 polyclonal sera and Pdmx-NT1 clinical sera (Fig. 5E). Furthermore, the synthetic peptide RVLAPALDSWGT competed for the binding of anti-DP1 antibodies present in sera of Pdmx-NT1 diseased (Fig. 5E). Data combined from Western blot and immunocytochemistry analysis suggested that peptide RVLAPALDSWGT could embed a structural as well as a linear epitope given that upon competition it interfered with DP1-specific serorecognition of globular as well as denatured epitopes (Fig. 5D and E).
Fig. 5.
Validation of DP1 as a true antigenic target in NT1 disease.
A. Human DP1 is expressed widely in different brain regions, and by mesenchymal and cancer stem cells. Cerebral cortex: Frontal cerebral cortex, Occipital lobe, Frontal lobe, Parietal lobe, Hippocampus; brain nuclei: Tectum, Amygdala, Thalamus, Caudate nucleus, Substantia nigra, Putamen, Nucleus ruber, Globus pallidus; Cerebellum: Cerebellar nuclei, Cerebellum right, Cerebellum left; Brainstem: Ventral pons, Dorsal pons, Dorsal medulla, Ventral medulla; Axonal tracts: Infundibulum, Optical nerve, Dorsal caudal pedunculi, Pedunculi, Cerebellum white matter, Corpus callosum; human glioma cells (hGC_1, hGC_2); human mesenchymal stem cells (hMSC); NC – negative control. PCR analyses were done using specific primers for human DP1. GAPDH mRNA expression was used to normalize the data across samples.
B. PGD2/DP1 signaling is associated with inflammation regulation. Expressions of human DP1, IL-1β, MMP1, IFNγ and IDO-1 were analyzed by PCR in human mesenhymal stem cells treated with PGD2 (10 μM) or cytokines IL-1β (1 ng/mL) and IFNγ (2 ng/mL). NC – negative control. GAPDH mRNA expression was used to normalize the data across samples.
C. RVLAPALD was identified as a target antigen sequence for NT1-specific polyclonal IgG response. Western blot analysis of phage particles containing the RVLAPALD-pIII (phage 1) or FLAG-pIII fusion proteins (phage 2) show that human Pdmx-NT1 serum (dilution 1:500) reacted specifically with the pIII protein containing the peptide RVLAPALDSWGT sequence, but not with the phage backbone or FLAG-pIII fusion protein. Duplicate membranes were incubated with Pdmx-NT1 sera treated with the synthetic peptide (RVLAPALDSWGTGGGDYKDDD: final conc 6.6 μg/mL) that significantly blocked the interaction between phage #1 and human IgG similarly to anti-FLAG antibody (dilution 1:2000) and phage #2. RVLAPALDSWGT-pIII fusion protein was not detected by HC sera (1: 500). Protein size markers are indicated at the right side of blot.
D. NT1-specific seroreactivity to DP1 protein is specifically blocked by RVLAPALD peptide. Western blot analysis of endogenous levels of DP1 protein (MW 40 kDa) in human glioma hGC_1 and hGC_2 cells using anti-PTGDR1 (DP1) polyclonal antibodies (1:500) (left, first panels). Note that, hGC_2 cells were negative of DP1 expression. The use of the Pdmx-NT1 serum (1:500) showed similar pattern of DP1 reactivity in hGC_1 and hGC_2 cells, and the DP1-specific signal was attenuated by pre-treatments of Pdmx-NT1 sera with a synthetic peptide #1 (final conc 6.6 μg/mL). Anti-GAPDH monoclonal antibody (1:10,000) was used as a control for immunoblots.
E. DP1 expressed by hMSCs and cancer was specifically blocked by RVLAPALD peptide. IF analysis of DP1 in hMSC and glioma cells. The antibodies used included: anti-PTGDR1 (1:500; green), Pdmx-NT1 serum (1:400; red) and the secondary Alexa Flour 488 and 647 (Invitrogen, 1:2000) antibodies. For antibody-blocking, Pdmx-NT1 sera (1:400) and synthetic peptide #1 (final conc 6.6 μg/mL) were used. Cells were analyzed for phalloidin-labelled cytoskeleton proteins (green, left) and nuclear structures (Hoechst 33342, blue). Abbreviations: Pdmx-NT1- Pandermix-induced narcolepsy type 1; HC – healthy control.
Fig. 6.
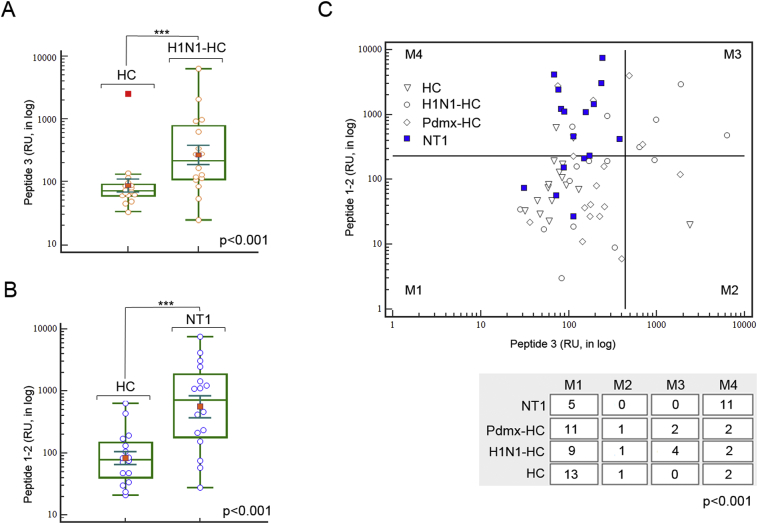
Use of MVA-defined peptides for immunodiagnostic applications of NT1. A. ELISPOT analysis of peptides containing H1N1/HA-specific sequence ESTRYQL (peptide_3) confirmed the power of the peptide to discriminate samples of natural H1N1 infection from HC (ANOVA p < .001). In box plots - yellow circles mark the single individuals of study cohorts; red dots mark the mean values; line marks the median values; inner whiskers mark confidence intervals for the mean; boxes mark the upper and lower quartiles; outer whiskers mark the max and the min values (excluding outliers).
B. ELISPOT analysis of peptides containing RVLAPALD (epitope of DP1) and KAPSAS (epitope of BCL6) (peptide_1 and _2, respectively) resulted in correct assignment of 10 Pdmx-NT1 samples and 6 sNT1 samples to NT1 group (ANOVA p < .001). On box plots - blue circles mark the single individuals of study cohorts; red dots mark the mean values; line marks the median values; inner whiskers mark confidence intervals for the mean; boxes mark the upper and lower quartiles; outer whiskers mark the max and the min values (excluding outliers).
C. ELISPOT analysis data combined revealed the power of peptides 1, 2, and 3 to discriminate 11 (7 Pdmx-NT1 and 4 sNT1) out of 16 NT1 samples across all samples (p < .001 Chi-squared test). Thresholds were calculated by using ROC curve analysis and results were visualised using scatter plot analysis with divided threshold values (M1-M4). Statistical significance of differences was calculated by two-way classification Chi-square test (Chi-squared p-value > .001). Abbreviations: HC– healthy control; H1N1-HC – H1N1 infected; Pdmx-HC- Pandemrix-vaccinated; NT1- narcolepsy type 1 (including 10 Pandemrix-induced NT1 samples).
We next studied whether the peptides identified here could be developed to an ELISPOT assay to discriminate sera in different disease groups. ELISPOT analysis data showed that peptides containing H1N1/HA-specific sequence ESTRYQL (peptide_3) discriminated between naturally H1N1 infected and healthy samples with no earlier H1N1 infection (ANOVA p < .001, Fig. 6A). RVLAPALD (epitope on DP1) and KAPSAS (epitope on BCL6) (peptide_1 and _2) peptides that were selected upon MVA data, correctly assigned upon ELISPOT analysis the NT1 group from HC samples (ANOVA p < .001, Fig. 6B). Combined ELISPOT analysis using all 3 peptides, could correctly classify 11 out of 16 NT1 (specifically - 7 Pdmx-NT1 and 3 NT1) samples across all controls (p < .001 Chi-squared test, Fig. 6C). Notably, majority of the DP1 and BCL6-peptide-positive NT1 samples had undetectable OX findings from the related CSFs (with average values of 6.1 pg/mL), whereas those 4 that were negative by our ELISPOT measurements, had OX levels in respective CSF samples still low but in detectable range (with average values of 77 pg/mL (Fig. 6C and see Materials and methods). Unlike the IgG response, the IgM levels in response to the tested peptides were low or absent in all studied individuals (data not shown). These findings confirmed that peptides carrying the epitope motifs identified in the study could be used in ELISPOT analysis to develop a novel multi-biomarker diagnostic assay for NT1.
4. Discussion
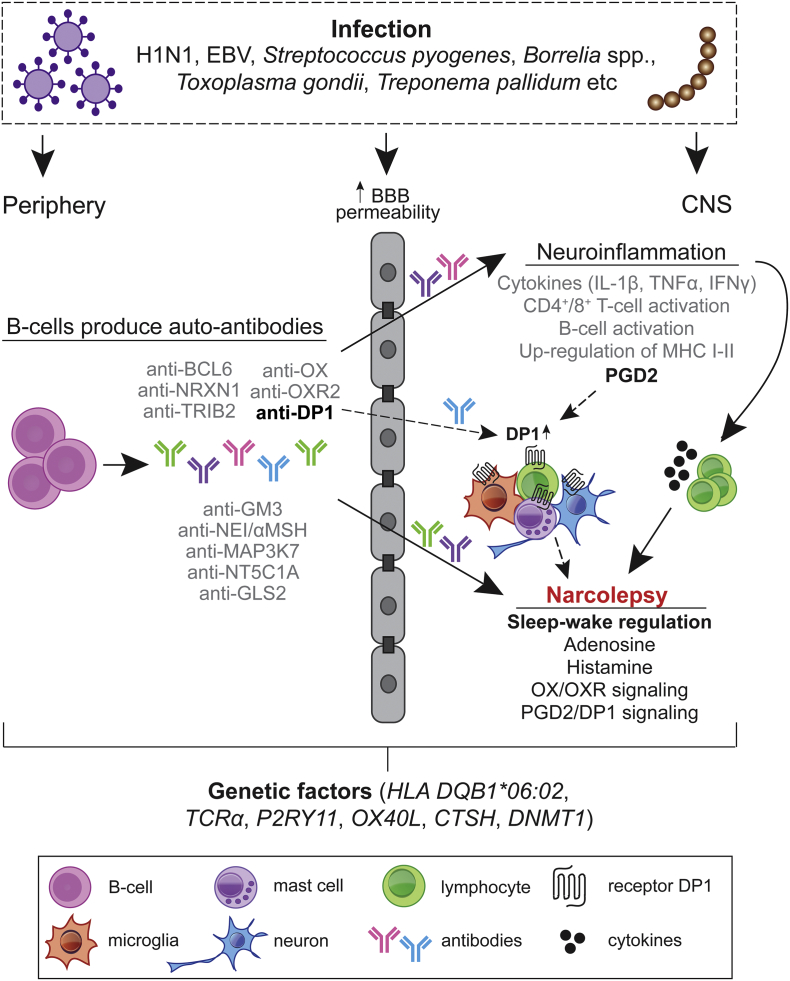
Despite extensive research using biomarker and neurophysiological approaches, known heterogeneity among NT1 diseased is not always consistent with serologic marker-based subtype classification schemes. Using an unbiased analysis of serum samples from single individuals, we detected a high variance in humoral immune response profiles, both in healthy and diseased people. We found that variance in immunoprofiles representing multifactorial heterogeneity of NT1 clearly determined distinct disease-specific serological profiles. We focused our analysis on peptides specific to Pdmx-immunized and -NT1 diseased subjects, which encompassed vaccine antigens and autoantigens in order to have a full coverage of potential triggers of the disease. Our results show that patients with NT1 exhibit a specific immune response to epitopes of receptor DP1. This finding highlights the importance of the PGD2-DP1 pathway in the functioning of sleep-wake homeostasis as suggested by the role of DP1 in slow-wave sleep (Terao et al., 1998). However, the precise mechanism by which PGD2-DP1 signaling may influence orexinergic neurons and immune regulation in NT1 requires further studies. In addition, using MVA-based immunoprofiling, we discovered epitopes, such as those of the protein BCL6 specific for Pdmx-NT1 and sNT1 patient group. This underscores the complexity of NT1 with different molecular targets and pathways involved and contributing to the immune response. Enhanced inflammation due to immune system malfunction has been detected in human narcoleptics in the regions of OX cell loss (Bassetti et al., 2010; John et al., 2013; Nishino, 2011; Thannickal et al., 2000, 2003, 2009), but the exact factors or mediators leading to the ultimate death of OX neurons are yet unknown.
Here we show that the DP1 receptor is linked to NT1 by acting as a possible antigen in the disease process. Prostaglandins play a key role in the inflammatory response and their synthesis is significantly increased after tissue injury and cell stress (Ricciotti and Fitzgerald, 2011). PGD2 is a major eicosanoid both in the Central Nervous System (CNS) and peripheral tissues with a role in inflammation as well as homeostasis (Jowsey et al., 2001; Vijay et al., 2017). PGD2 is abundantly produced by mast cells and Th2 cells, and among a wide range of other body cells (see ref. in Farhat et al. (2011)). PGD2 elicits its downstream effects by activating DP1 and DP2 receptors with opposing effects on cyclic AMP (cAMP) production, and/or phosphoinositol turnover and intracellular Ca2+ mobilization (Liang et al., 2005). In the brain, PGD2 regulates sleep, body temperature, and nociception and its levels exhibit marked changes in different neuropathologies (reviewed in (Liang et al., 2005, Mohri et al., 2006, Urade and Hayaishi, 2011)). The microglial PGD2-DP1 pathway is also known to mediate neuronal damage through microglial activation (Bate et al., 2006; Vijay et al., 2017).
Among cells expressing DP1, mast cells (MCs) can release histamine and other factors that affect sleep and the immune response in the brain. Accumulating evidence shows that MCs play a role in the regulation of sleep and behavior (Chikahisa et al., 2013). MCs are most abundant in young individuals under the age of 19, after which their counts decline with age (Porzionato et al., 2004; Turygin et al., 2005). Most significantly, the maturation of MCs is influenced by PGD2 and the receptor DP1 (Taketomi et al., 2013). In addition to histamine, DP1 signaling may also influence the levels of adenosine that is known to regulate sleep (Urade and Hayaishi, 2011). The precise role of MCs and their released factors such as histamine in NT1 warrants further studies.
In conclusion, the present study shows that anti-DP1 antibodies are autoimmune agents in the course of NT1 prompting more studies on the role of PDG2-DP1 signaling in OX-signaling and in the disease. Currently DP1-selective agonist/antagonist therapies are considered in treating autoimmune disorders such as asthma (Maicas et al., 2012; Santini et al., 2016; Santus and Radovanovic, 2016). Our data also indicate that, depending on the antibody concentrations and affinities, anti-DP1 antibodies may modify the function of pharmaceutical compounds targeting PGD2-DP1–signaling pathways (Narumiya and Fitzgerald, 2001) that need to be taken into account in clinical studies.
Supporting evidence that the humoral response in the CNS is derived from different peripheral tissue antigens is provided by the findings that sera from NT1 diseased can bind brain and muscle structures (Ahmed et al., 2014; Smith et al., 2004). There is a plethora of data that genetic or experimental alterations of the OX system are associated with NT1, however, OXs are not restricted to the CNS and together with their receptors OX1R and OX2R are widely expressed in peripheral tissues (see ref. in (Voisin et al., 2003)). BCL6, another NT1 antigen, is a master regulator required in mature B-cells during germinal center (GC) reaction (Ref in Pei et al. (2017)). NRXN1-α has been isolated from brain and heart tissues suggesting a role also in heart development (Nagase et al., 1998). TRIB2 is present in many cell populations both in and outside the nervous system, including the immune cells (Eder et al., 2008; Sung et al., 2006). Gangliosides (anti-GM3) are abundant in the brain, but in extraneural tissues, relatively high concentrations of ganglio-series GMs were found in bone marrow, erythrocytes, intestine, liver, spleen, testis, kidney, and in embryonic stem cells (Kolter, 2012). NEI-MCH has mostly been detected in peripheral organs (Viale et al., 1997). NT5C1A is highly abundant in skeletal muscle tissue (Hunsucker et al., 2001). GLS2 is expressed specifically in the liver, but also in extrahepatic tissues, like the brain, pancreas, cells of the immune system (ref in Martin-Rufian et al. (2012)). However, it remains elusive what pathogenic roles these antibodies against the above-mentioned proteins may exert within the periphery.
Our data of immunoprofiling support the existence of immune defects in multiple pathways associating NT1 to a) DP1 and PDG2/histamine associated disorders, b) BCL6 and the chronic status of latent herpesviruses (such as EBV), c) orexin/OX1/2R–related dysfunctions, d) stress and inflammation-associated mitogen-activated pathways (such as MAP3K7, also known as transforming growth factor (TGF)-β-activated kinase 1 (TAK1)), and e) adenosine-deficiency linked dysfunctions (involving NT5C1A) (Table S1, Fig. 7). Together these results provide a comprehensive map of potential molecular targets contributing to NT1 that can be of help in designing future strategies for the diagnostics and treatment of the disease. More broadly, our study demonstrates the usefulness of MVA as a method for disease classification and for the discovery of novel biomarkers that can be applicable to any human disease.
Fig. 7.
Hypothetical model for the aggravation of autoimmune response in Pdmx-associated and spontaneous NT1.
The immune response in NT1 is highly heterogenic with different pathways affected during the disease progression. We favor the idea that the lifelong risk for NT1 or for disease aggravation in pre-disposed individuals is increased following inflammatory triggers upon breaching of the blood–brain barrier (BBB) and with activation of preexisting auto-reactive antibodies (Ab) and cells reaching brain. Consequently, an immune response to A/H1N1 (and subsequent molecular mimicry) or a generalized stimulation of the immune system mediated by the Pdmx vaccine as AS03-adjuvanted vaccine can act as the inflammatory trigger (Morel et al., 2011; Carmona et al., 2010; Meyer et al., 2011). The inflammation triggers include i) infections (examples of pathogens are shown), ii) genetic factors, or iii) chronic inflammation (Kornum et al., 2011). The polyclonal Ab response from peripheral tissue may initiate disease by concentrating antigens in the brain to presentation-competent cells (Getahun et al., 2004). Recent data show further that peripherally produced human anti-CNS reactive antibodies are capable of opsonizing human CNS antigens (Kinzel et al., 2016). The entry of immune cells (T cells, B cells, macrophages, microglia and mast cells) cause neuroinflammation with the release of cytokines that damage neurons including HCRT+ neurons involved in sleep/wake regulation. Production of auto-reactive antibodies as a secondary response to cell death of HCRT-or other brain-resident cells can occur via antigen presenting cells. Prostaglandins are part of the inflammatory response in the brain acting via specific receptors. In particular, DP1 is produced by astrocytes, oligodendrocytes, neurons, microglia and meningeal cells (Liang et al., 2005; Mohri et al., 2007; Beuckmann et al., 2000). PGD2 signaling is known to prevent excessive inflammasome activation and may act as an anti-inflammatory pathway in the brain. Additionally, in brain residing mast cells, DP1 activity promotes maturation and histamine release (Taketomi et al., 2013). The latter is of particular interest given that histamine levels in the CSF of NT1-diseased are reduced (Nishino et al., 2009). Thus, our findings suggest that the anti-DP1 immune response, whether causal or sequel, can interfere with PGD2 signaling in the brain. The results provide also evidence that the dysfunctional DP1 network can be a target for diagnosis and intervention of NT1, a conclusion that warrants further investigations.
Funding Sources
The research leading to these results was mainly funded by Protobios's institutional research funding from the Estonian Research Council (contract no. 5.1-4/1373). KP was partially supported by the institutional research grant IUT19-18 of TUT from the Estonian Research Council. MP was partially supported by NARPA funding from the Academy of Finland (grant no. 260603), Sigrid Jusélius Foundation and Magnus Ehrnrooth Foundation. The EU Research and Innovation Staff Exchange (RISE) program (SZ_TEST H2020-MSCA-RISE-2016 (EU734791)) provided additional funding. The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the manuscript.
Conflicts of Interest
AV, OV, MP, KP, TN, and KP are co-applicants on a pending patent application related to diagnosing of narcolepsy (WO 2017/203106). All other authors declare no competing interests.
Author Contributions
HS, AP, TN, MP, AV, and KP contributed to the design of the study. MP, OV, and AV were members of NARPANord consortium, MP was the chairman. HS, AP, AK, SP, MJ, MP, and AV contributed to data collection. HS, AP, AK, SP, MJ, DL, and KP contributed to the development and implementation of the data analysis plan. JV and PA were responsible for data management and pattern recognition analysis of the MVA data. All authors were involved in data interpretation, drafting, review, and approval of the report, and the decision to submit for publication.
Acknowledgements
Our deep gratitude goes to Lagle Kasak, Urmas Liivas, Grete Rullinkov, Veronika Tammekivi, Helen Verev, Mirjam Luhakooder, and Maila Rähn (Protobios, Estonia) as well as to Anne Huutoniemi, Kirsi Aaltonen, Sanna Mäki, and Mira Utriainen (UH, Finland).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.01.043.
Appendix A. Supplementary Data
Supplementary material
References
- Ahmed S.S., Schur P.H., Macdonald N.E., Steinman L. Narcolepsy, 2009 A(H1N1) pandemic influenza, and pandemic influenza vaccinations: what is known and unknown about the neurological disorder, the role for autoimmunity, and vaccine adjuvants. J. Autoimmun. 2014;50:1–11. doi: 10.1016/j.jaut.2014.01.033. [DOI] [PubMed] [Google Scholar]
- Ahmed S.S., Volkmuth W., Duca J., Corti L., Pallaoro M., Pezzicoli A., Karle A., Rigat F., Rappuoli R., Narasimhan V., Julkunen I., Vuorela A., Vaarala O., Nohynek H., Pasini F.L., Montomoli E., Trombetta C., Adams C.M., Rothbard J., Steinman L. Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2. Sci. Transl. Med. 2015;7:294ra105. doi: 10.1126/scitranslmed.aab2354. [DOI] [PubMed] [Google Scholar]
- Anastasina M., Domanska A., Palm K., Butcher S. Human picornaviruses associated with neurological diseases and their neutralization by antibodies. J. Gen. Virol. 2017;98:1145–1158. doi: 10.1099/jgv.0.000780. [DOI] [PubMed] [Google Scholar]
- Aran A., Lin L., Nevsimalova S., Plazzi G., Hong S.C., Weiner K., Zeitzer J., Mignot E. Elevated anti-streptococcal antibodies in patients with recent narcolepsy onset. Sleep. 2009;32:979–983. doi: 10.1093/sleep/32.8.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avlani V.A., Gregory K.J., Morton C.J., Parker M.W., Sexton P.M., Christopoulos A. Critical role for the second extracellular loop in the binding of both orthosteric and allosteric G protein-coupled receptor ligands. J. Biol. Chem. 2007;282:25677–25686. doi: 10.1074/jbc.M702311200. [DOI] [PubMed] [Google Scholar]
- Ayoglu B., Schwenk J.M., Nilsson P. Antigen arrays for profiling autoantibody repertoires. Bioanalysis. 2016;8:1105–1126. doi: 10.4155/bio.16.31. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bassetti C.L., Baumann C.R., Dauvilliers Y., Croyal M., Robert P., Schwartz J.C. Cerebrospinal fluid histamine levels are decreased in patients with narcolepsy and excessive daytime sleepiness of other origin. J. Sleep Res. 2010;19:620–623. doi: 10.1111/j.1365-2869.2010.00819.x. [DOI] [PubMed] [Google Scholar]
- Bate C., Kempster S., Williams A. Prostaglandin D2 mediates neuronal damage by amyloid-beta or prions which activates microglial cells. Neuropharmacology. 2006;50:229–237. doi: 10.1016/j.neuropharm.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Bergman P., Adori C., Vas S., Kai-Larsen Y., Sarkanen T., Cederlund A., Agerberth B., Julkunen I., Horvath B., Kostyalik D., Kalmar L., Bagdy G., Huutoniemi A., Partinen M., Hokfelt T. Narcolepsy patients have antibodies that stain distinct cell populations in rat brain and influence sleep patterns. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3735–44. doi: 10.1073/pnas.1412189111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuckmann C.T., Lazarus M., Gerashchenko D., Mizoguchi A., Nomura S., Mohri I., Uesugi A., Kaneko T., Mizuno N., Hayaishi O., Urade Y. Cellular localization of lipocalin-type prostaglandin D synthase (beta-trace) in the central nervous system of the adult rat. J. Comp. Neurol. 2000;428:62–78. doi: 10.1002/1096-9861(20001204)428:1<62::aid-cne6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Bomfim I.L., Lamb F., Fink K., Szakacs A., Silveira A., Franzen L., Azhary V., Maeurer M., Feltelius N., Darin N., Hallbook T., Arnheim-Dahlstrom L., Kockum I., Olsson T. The immunogenetics of narcolepsy associated with A(H1N1)pdm09 vaccination (Pandemrix) supports a potent gene-environment interaction. Genes Immun. 2017;18:75–81. doi: 10.1038/gene.2017.1. [DOI] [PubMed] [Google Scholar]
- Bonvalet M., Ollila H.M., Ambati A., Mignot E. Autoimmunity in narcolepsy. Curr. Opin. Pulm. Med. 2017;23:522–529. doi: 10.1097/MCP.0000000000000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A., Jonassen I., Vilo J., Ukkonen E. Predicting gene regulatory elements in silico on a genomic scale. Genome Res. 1998;8:1202–1215. doi: 10.1101/gr.8.11.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona A., Omenaca F., Tejedor J.C., Merino J.M., Vaman T., Dieussaert I., Gillard P., Aristegui J. Immunogenicity and safety of AS03-adjuvanted 2009 influenza A H1N1 vaccine in children 6-35 months. Vaccine. 2010;28:5837–5844. doi: 10.1016/j.vaccine.2010.06.065. [DOI] [PubMed] [Google Scholar]
- Chikahisa S., Kodama T., Soya A., Sagawa Y., Ishimaru Y., Sei H., Nishino S. Histamine from brain resident MAST cells promotes wakefulness and modulates behavioral states. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen A., Kringelum J.V., Hansen C.S., Bogh K.L., Sullivan E., Patel J., Rigby N.M., Eiwegger T., Szepfalusi Z., DE Masi F., Nielsen M., Lund O., Dufva M. High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum. Sci. Rep. 2015;5:12913. doi: 10.1038/srep12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic-Lopes V., Bayer L., Dorsaz S., Maret S., Pradervand S., Dauvilliers Y., Lecendreux M., Lammers G.J., Donjacour C.E., DU Pasquier R.A., Pfister C., Petit B., Hor H., Muhlethaler M., Tafti M. Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J. Clin. Invest. 2010;120:713–719. doi: 10.1172/JCI41366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Herran-Arita A.K., Kornum B.R., Mahlios J., Jiang W., Lin L., Hou T., Macaubas C., Einen M., Plazzi G., Crowe C., Newell E.W., Davis M.M., Mellins E.D., Mignot E. CD4+ T cell autoimmunity to hypocretin/orexin and cross-reactivity to a 2009 H1N1 influenza A epitope in narcolepsy. Sci. Transl. Med. 2013;5:216ra176. doi: 10.1126/scitranslmed.3007762. (retracted) [DOI] [PubMed] [Google Scholar]
- Eder K., Guan H., Sung H.Y., Ward J., Angyal A., Janas M., Sarmay G., Duda E., Turner M., Dower S.K., Francis S.E., Crossman D.C., Kiss-Toth E. Tribbles-2 is a novel regulator of inflammatory activation of monocytes. Int. Immunol. 2008;20:1543–1550. doi: 10.1093/intimm/dxn116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency . Pandemrix: European Public Assessment Report. 2009. Annex I Summary of Product Characteristics; pp. 1–20. (first published) [Google Scholar]
- Farhat A., Philibert P., Sultan C., Poulat F., Boizet-Bonhoure B. Hematopoietic-prostaglandin D2 synthase through PGD2 production is involved in the adult ovarian physiology. J. Ovarian Res. 2011;4:3. doi: 10.1186/1757-2215-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getahun A., Dahlström J., Wernersson S., Heyman B. IgG2a-mediated enhancement of antibody and T cell responses and its relation to inhibitory and activating Fc gamma receptors. J. Immunol. 2004;172:5269–5276. doi: 10.4049/jimmunol.172.9.5269. [DOI] [PubMed] [Google Scholar]
- Gianfrani C., Oseroff C., Sidney J., Chesnut R.W., Sette A. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum. Immunol. 2000;61:438–452. doi: 10.1016/s0198-8859(00)00105-1. [DOI] [PubMed] [Google Scholar]
- Haggmark-Manberg A., Zandian A., Forsstrom B., Khademi M., Lima Bomfim I., Hellstrom C., Arnheim-Dahlstrom L., Hallbook T., Darin N., Lundberg I.E., Uhlen M., Partinen M., Schwenk J.M., Olsson T., Nilsson P. Autoantibody targets in vaccine-associated narcolepsy. Autoimmunity. 2016;49:421–433. doi: 10.1080/08916934.2016.1183655. [DOI] [PubMed] [Google Scholar]
- Hallmayer J., Faraco J., Lin L., Hesselson S., Winkelmann J., Kawashima M., Mayer G., Plazzi G., Nevsimalova S., Bourgin P., Hong S.C., Honda Y., Honda M., Högl B., Longstreth W.T.J., Montplaisir J., Kemlink D., Einen M., Chen J., Musone S.L., Akana M., Miyagawa T., Duan J., Desautels A., Erhardt C., Hesla P.E., Poli F., Frauscher B., Jeong J.H., Lee S.P., Ton T.G., Kvale M., Kolesar L., Dobrovolná M., Nepom G.T., Salomon D., Wichmann H.E., Rouleau G.A., Gieger C., Levinson D.F., Gejman P.V., Meitinger T., Young T., Peppard P., Tokunaga K., Kwok P.Y., Risch N., Mignot E. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat. Genet. 2009;41:708–711. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Lin L., Warby S.C., Faraco J., Li J., Dong S.X., An P., Zhao L., Wang L.H., Li Q.Y., Yan H., Gao Z.C., Yuan Y., Strohl K.P., Mignot E. Narcolepsy onset is seasonal and increased following the 2009 H1N1 pandemic in China. Ann. Neurol. 2011;70:410–417. doi: 10.1002/ana.22587. [DOI] [PubMed] [Google Scholar]
- Han F., Lin L., Li J., Dong X.S., Mignot E. Decreased incidence of childhood narcolepsy 2 years after the 2009 H1N1 winter flu pandemic. Ann. Neurol. 2013;73:560. doi: 10.1002/ana.23799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Lin L., Schormair B., Pizza F., Plazzi G., Ollila H.M., Nevsimalova S., Jennum P., Knudsen S., Winkelmann J., Coquillard C., Babrzadeh F., Strom T.M., Wang C., Mindrinos M., Fernandez Vina, M. & Mignot, E. HLA DQB1*06:02 negative narcolepsy with hypocretin/orexin deficiency. Sleep. 2014;37:1601–1608. doi: 10.5665/sleep.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsucker S.A., Spychala J., Mitchell B.S. Human cytosolic 5′-nucleotidase I: characterization and role in nucleoside analog resistance. J. Biol. Chem. 2001;276:10498–10504. doi: 10.1074/jbc.M011218200. [DOI] [PubMed] [Google Scholar]
- Ionov Y. A high throughput method for identifying personalized tumor-associated antigens. Oncotarget. 2010;1:148–155. doi: 10.18632/oncotarget.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Zeisel A., Joost S., La Manno G., Zajac P., Kasper M., Lönnerberg P., Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- Jaager K., Neuman T. Human dermal fibroblasts exhibit delayed adipogenic differentiation compared with mesenchymal stem cells. Stem Cells Dev. 2011;20:1327–1336. doi: 10.1089/scd.2010.0258. [DOI] [PubMed] [Google Scholar]
- Jacob L., Leib R., Ollila H.M., Bonvalet M., Adams C.M., Mignot E. Comparison of Pandemrix and Arepanrix, two pH1N1 AS03-adjuvanted vaccines differentially associated with narcolepsy development. Brain Behav. Immun. 2015;47:44–57. doi: 10.1016/j.bbi.2014.11.004. [DOI] [PubMed] [Google Scholar]
- John J., Thannickal T.C., Mcgregor R., Ramanathan L., Ohtsu H., Nishino S., Sakai N., Yamanaka A., Stone C., Cornford M., Siegel J.M. Greatly increased numbers of histamine cells in human narcolepsy with cataplexy. Ann. Neurol. 2013;74:786–793. doi: 10.1002/ana.23968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowsey I.R., Thomson A.M., Flanagan J.U., Murdock P.R., Moore G.B., Meyer D.J., Murphy G.J., Smith S.A., Hayes J.D. Mammalian class Sigma glutathione S-transferases: catalytic properties and tissue-specific expression of human and rat GSH-dependent prostaglandin D2 synthases. Biochem. J. 2001;359:507–516. doi: 10.1042/0264-6021:3590507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzav A., Arango M.T., Kivity S., Tanaka S., Givaty G., Agmon-Levin N., Honda M., Anaya J.M., Chapman J., Shoenfeld Y. Passive transfer of narcolepsy: anti-TRIB2 autoantibody positive patient IgG causes hypothalamic orexin neuron loss and sleep attacks in mice. J. Autoimmun. 2013;45:24–30. doi: 10.1016/j.jaut.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Kauts M.L., Pihelgas S., Orro K., Neuman T., Piirsoo A. CCL5/CCR1 axis regulates multipotency of human adipose tissue derived stromal cells. Stem Cell Res. 2013;10:166–178. doi: 10.1016/j.scr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Kinzel S., Lehmann-Horn K., Torke S., Hausler D., Winkler A., Stadelmann C., Payne N., Feldmann L., Saiz A., Reindl M., Lalive P.H., Bernard C.C., Bruck W., Weber M.S. Myelin-reactive antibodies initiate T cell-mediated CNS autoimmune disease by opsonization of endogenous antigen. Acta Neuropathol. 2016;132:43–58. doi: 10.1007/s00401-016-1559-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S., Biering-Sorensen B., Kornum B.R., Petersen E.R., Ibsen J.D., Gammeltoft S., Mignot E., Jennum P.J. Early IVIg treatment has no effect on post-H1N1 narcolepsy phenotype or hypocretin deficiency. Neurology. 2012;79:102–103. doi: 10.1212/WNL.0b013e31825dce03. [DOI] [PubMed] [Google Scholar]
- Kolter T. Ganglioside biochemistry. ISRN Biochem. 2012;2012:506160. doi: 10.5402/2012/506160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornum B.R., Faraco J., Mignot E. Narcolepsy with hypocretin/orexin deficiency, infections and autoimmunity of the brain. Curr. Opin. Neurobiol. 2011;21:897–903. doi: 10.1016/j.conb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Kruup, M. (2013) Finding motifs from short peptides.
- Lecendreux M., Libri V., Jaussent I., Mottez E., Lopez R., Lavault S., Regnault A., Arnulf I., Dauvilliers Y. Impact of cytokine in type 1 narcolepsy: role of pandemic H1N1 vaccination? J. Autoimmun. 2015;60:20–31. doi: 10.1016/j.jaut.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Liang X., Wu L., Hand T., Andreasson K. Prostaglandin D2 mediates neuronal protection via the DP1 receptor. J. Neurochem. 2005;92:477–486. doi: 10.1111/j.1471-4159.2004.02870.x. [DOI] [PubMed] [Google Scholar]
- Lind A., Ramelius A., Olsson T., Arnheim-Dahlstrom L., Lamb F., Khademi M., Ambati A., Maeurer M., Nilsson A.L., Bomfim I.L., Fink K., Lernmark A. A/H1N1 antibodies and TRIB2 autoantibodies in narcolepsy patients diagnosed in conjunction with the Pandemrix vaccination campaign in Sweden 2009-2010. J. Autoimmun. 2014;50:99–106. doi: 10.1016/j.jaut.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Longstreth W.T., Jr., Ton T.G., Koepsell T.D. Narcolepsy and streptococcal infections. Sleep. 2009;32:1548. doi: 10.1093/sleep/32.12.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicas N., Ibanez L., Alcaraz M.J., Ubeda A., Ferrandiz M.L. Prostaglandin D2 regulates joint inflammation and destruction in murine collagen-induced arthritis. Arthritis Rheum. 2012;64:130–140. doi: 10.1002/art.30656. [DOI] [PubMed] [Google Scholar]
- Martin-Rufian M., Tosina M., Campos-Sandoval J.A., Manzanares E., Lobo C., Segura J.A., Alonso F.J., Mates J.M., Marquez J. Mammalian glutaminase Gls2 gene encodes two functional alternative transcripts by a surrogate promoter usage mechanism. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Adam M., Schweiger B., Ilchmann C., Eulenburg C., Sattinger E., Runte H., Schluter M., Deuse T., Reichenspurner H., Costard-Jackle A. Antibody response after a single dose of an AS03-adjuvanted split-virion influenza A (H1N1) vaccine in heart transplant recipients. Transplantation. 2011;91:1031–1035. doi: 10.1097/TP.0b013e3182115be0. [DOI] [PubMed] [Google Scholar]
- Mohri I., Taniike M., Taniguchi H., Kanekiyo T., Aritake K., Inui T., Fukumoto N., Eguchi N., Kushi A., Sasai H., Kanaoka Y., Ozono K., Narumiya S., Suzuki K., Urade Y. Prostaglandin D2-mediated microglia/astrocyte interaction enhances astrogliosis and demyelination in twitcher. J. Neurosci. 2006;26:4383–4393. doi: 10.1523/JNEUROSCI.4531-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri I., Kadoyama K., Kanekiyo T., Sato Y., Kagitani-Shimono K., Saito Y., Suzuki K., Kudo T., Takeda M., Urade Y., Murayama S., Taniike M. Hematopoietic prostaglandin D synthase and DP1 receptor are selectively upregulated in microglia and astrocytes within senile plaques from human patients and in a mouse model of Alzheimer disease. J. Neuropathol. Exp. Neurol. 2007;66:469–480. doi: 10.1097/01.jnen.0000240472.43038.27. [DOI] [PubMed] [Google Scholar]
- Morel S., Didierlaurent A., Bourguignon P., Delhaye S., Baras B., Jacob V., Planty C., Elouahabi A., Harvengt P., Carlsen H., Kielland A., Chomez P., Garcon N., VAN Mechelen M. Adjuvant System AS03 containing alpha-tocopherol modulates innate immune response and leads to improved adaptive immunity. Vaccine. 2011;29:2461–2473. doi: 10.1016/j.vaccine.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Nagase T., Ishikawa K., Suyama M., Kikuno R., Hirosawa M., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998;5:355–364. doi: 10.1093/dnares/5.6.355. [DOI] [PubMed] [Google Scholar]
- Nagata N., Iwanari H., Kumagai H., Kusano-Arai O., Ikeda Y., Aritake K., Hamakubo T., Urade Y. Generation and characterization of an antagonistic monoclonal antibody against an extracellular domain of mouse DP2 (CRTH2/GPR44) receptors for prostaglandin D2. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumiya S., Fitzgerald G.A. Genetic and pharmacological analysis of prostanoid receptor function. J. Clin. Invest. 2001;108:25–30. doi: 10.1172/JCI13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson C., Harvey R., Johnson R., Guilfoyle K., Engelhardt O.G., Robertson J.S. An additional oligosaccharide moiety in the HA of a pandemic influenza H1N1 candidate vaccine virus confers increased antigen yield in eggs. Vaccine. 2012;30:745–751. doi: 10.1016/j.vaccine.2011.11.081. [DOI] [PubMed] [Google Scholar]
- Nishino S. Hypothalamus, hypocretins/orexin, and vigilance control. Handb. Clin. Neurol. 2011;99:765–782. doi: 10.1016/B978-0-444-52007-4.00006-0. [DOI] [PubMed] [Google Scholar]
- Nishino S., Sakurai E., Nevsimalova S., Yoshida Y., Watanabe T., Yanai K., Mignot E. Decreased CSF histamine in narcolepsy with and without low CSF hypocretin-1 in comparison to healthy controls. Sleep. 2009;32:175–180. doi: 10.1093/sleep/32.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partinen M., Kornum B.R., Plazzi G., Jennum P., Julkunen I., Vaarala O. Narcolepsy as an autoimmune disease: the role of H1N1 infection and vaccination. Lancet Neurol. 2014;13:600–613. doi: 10.1016/S1474-4422(14)70075-4. [DOI] [PubMed] [Google Scholar]
- Pei Y., Banerjee S., Jha H.C., Sun Z., Robertson E.S. An essential EBV latent antigen 3C binds Bcl6 for targeted degradation and cell proliferation. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyron C., Faraco J., Rogers W., Ripley B., Overeem S., Charnay Y., Nevsimalova S., Aldrich M., Reynolds D., Albin R., Li R., Hungs M., Pedrazzoli M., Padigaru M., Kucherlapati M., Fan J., Maki R., Lammers G.J., Bouras C., Kucherlapati R., Nishino S., Mignot E. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat. Med. 2000;6:991–997. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- Porzionato A., Macchi V., Parenti A., De Caro R. The distribution of mast cells in the human area postrema. J. Anat. 2004;204:141–147. doi: 10.1111/j.1469-7580.2004.00256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciotti E., Fitzgerald G.A. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J.S., Nicolson C., Harvey R., Johnson R., Major D., Guilfoyle K., Roseby S., Newman R., Collin R., Wallis C., Engelhardt O.G., Wood J.M., Le J., Manojkumar R., Pokorny B.A., Silverman J., Devis R., Bucher D., Verity E., Agius C., Camuglia S., Ong C., Rockman S., Curtis A., Schoofs P., Zoueva O., Xie H., Li X., Lin Z., Ye Z., Chen L.M., O'neill E., Balish A., Lipatov A.S., Guo Z., Isakova I., Davis C.T., Rivailler P., Gustin K.M., Belser J.A., Maines T.R., Tumpey T.M., Xu X., Katz J.M., Klimov A., Cox N.J., Donis R.O. The development of vaccine viruses against pandemic A(H1N1) influenza. Vaccine. 2011;29:1836–1843. doi: 10.1016/j.vaccine.2010.12.044. [DOI] [PubMed] [Google Scholar]
- Santini G., Mores N., Malerba M., Mondino C., Macis G., Montuschi P. Investigational prostaglandin D2 receptor antagonists for airway inflammation. Expert Opin. Investig. Drugs. 2016;25:639–652. doi: 10.1080/13543784.2016.1175434. [DOI] [PubMed] [Google Scholar]
- Santus P., Radovanovic D. Prostaglandin D2 receptor antagonists in early development as potential therapeutic options for asthma. Expert Opin. Investig. Drugs. 2016;25:1083–1092. doi: 10.1080/13543784.2016.1212838. [DOI] [PubMed] [Google Scholar]
- Sarkanen T., Alen R., Partinen M. Transient Impact of Rituximab in H1N1 Vaccination-associated Narcolepsy With Severe Psychiatric Symptoms. Neurologist. 2016;21:85–86. doi: 10.1097/NRL.0000000000000099. [DOI] [PubMed] [Google Scholar]
- Sarkanen T.O., Alakuijala A., Dauvilliers Y., Partinen M.M. Incidence of narcolepsy after H1N1 influenza and vaccinations: Systematic review and meta-analysis. Sleep Med. Rev. 2017 doi: 10.1016/j.smrv.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Schanen B.C., De Groot A.S., Moise L., Ardito M., Mcclaine E., Martin W., Wittman V., Warren W.L., Drake D.R., 3rd Coupling sensitive in vitro and in silico techniques to assess cross-reactive CD4(+) T cells against the swine-origin H1N1 influenza virus. Vaccine. 2011;29:3299–3309. doi: 10.1016/j.vaccine.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J., Jackson M.W., Neufing P., Mcevoy R.D., Gordon T.P. A functional autoantibody in narcolepsy. Lancet. 2004;364:2122–2124. doi: 10.1016/S0140-6736(04)17553-3. [DOI] [PubMed] [Google Scholar]
- Sung H.Y., Francis S.E., Crossman D.C., Kiss-Toth E. Regulation of expression and signalling modulator function of mammalian tribbles is cell-type specific. Immunol. Lett. 2006;104:171–177. doi: 10.1016/j.imlet.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Tafti M., Hor H., Dauvilliers Y., Lammers G.J., Overeem S., Mayer G., Javidi S., Iranzo A., Santamaria J., Peraita-Adrados R., Vicario J.L., Arnulf I., Plazzi G., Bayard S., Poli F., Pizza F., Geisler P., Wierzbicka A., Bassetti C.L., Mathis J., Lecendreux M., Donjacour C.E., Van Der Heide A., Heinzer R., Haba-Rubio J., Feketeova E., Hogl B., Frauscher B., Beneto A., Khatami R., Canellas F., Pfister C., Scholz S., Billiard M., Baumann C.R., Ercilla G., Verduijn W., Claas F.H., Dubois V., Nowak J., Eberhard H.P., Pradervand S., Hor C.N., Testi M., Tiercy J.M., Kutalik Z. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 2014;37:19–25. doi: 10.5665/sleep.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketomi Y., Ueno N., Kojima T., Sato H., Murase R., Yamamoto K., Tanaka S., Sakanaka M., Nakamura M., Nishito Y., Kawana M., Kambe N., Ikeda K., Taguchi R., Nakamizo S., Kabashima K., Gelb M.H., Arita M., Yokomizo T., Watanabe K., Hirai H., Okayama Y., Ra C., Aritake K., Urade Y., Morimoto K., Sugimoto Y., Shimizu T., Narumiya S., Hara S., Murakami M. Mast cell maturation is driven via a group III phospholipase A2-prostaglandin D2-DP1 receptor paracrine axis. Nat. Immunol. 2013;14:554–563. doi: 10.1038/ni.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao A., Matsumura H., Saito M. Interleukin-1 induces slow-wave sleep at the prostaglandin D2-sensitive sleep-promoting zone in the rat brain. J. Neurosci. 1998;18:6599–6607. doi: 10.1523/JNEUROSCI.18-16-06599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal T.C., Moore R.Y., Nienhuis R., Ramanathan L., Gulyani S., Aldrich M., Cornford M., Siegel J.M. Reduced number of hypocretin neurons in human narcolepsy. Neuron. 2000;27:469–474. doi: 10.1016/s0896-6273(00)00058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal T.C., Siegel J.M., Nienhuis R., Moore R.Y. Pattern of hypocretin (orexin) soma and axon loss, and gliosis, in human narcolepsy. Brain Pathol. 2003;13:340–351. doi: 10.1111/j.1750-3639.2003.tb00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal T.C., Nienhuis R., Siegel J.M. Localized loss of hypocretin (orexin) cells in narcolepsy without cataplexy. Sleep. 2009;32(8):993. doi: 10.1093/sleep/32.8.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turygin V.V., Babik T.M., Boyakov A.A. Characteristics of mast cells in the choroid plexus of the ventricles of the human brain in aging. Neurosci. Behav. Physiol. 2005;35:909–911. doi: 10.1007/s11055-005-0144-8. [DOI] [PubMed] [Google Scholar]
- Urade Y., Hayaishi O. Prostaglandin D2 and sleep/wake regulation. Sleep Med. Rev. 2011;15:411–418. doi: 10.1016/j.smrv.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Vaarala O., Vuorela A., Partinen M., Baumann M., Freitag T.L., Meri S., Saavalainen P., Jauhiainen M., Soliymani R., Kirjavainen T., Olsen P., Saarenpaa-Heikkila O., Rouvinen J., Roivainen M., Nohynek H., Jokinen J., Julkunen I., Kilpi T. Antigenic differences between AS03 adjuvanted influenza A (H1N1) pandemic vaccines: implications for pandemrix-associated narcolepsy risk. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko P.O., Gavrilov Y.V., Yamamoto M., Reddy H., Haybaeck J., Mignot E., Baumann C.R., Scammell T.E. Increase of histaminergic tuberomammillary neurons in narcolepsy. Ann. Neurol. 2013;74:794–804. doi: 10.1002/ana.24019. [DOI] [PubMed] [Google Scholar]
- Vassalli A., Li S., Tafti M. Comment on "Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2". Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aad2353. [DOI] [PubMed] [Google Scholar]
- Viale A., Zhixing Y., Breton C., Pedeutour F., Coquerel A., Jordan D., Nahon J.L. The melanin-concentrating hormone gene in human: flanking region analysis, fine chromosome mapping, and tissue-specific expression. Brain Res. Mol. Brain Res. 1997;46:243–255. doi: 10.1016/s0169-328x(97)00018-1. [DOI] [PubMed] [Google Scholar]
- Vijay R., Fehr A.R., Janowski A.M., Athmer J., Wheeler D.L., Grunewald M., Sompallae R., Kurup S.P., Meyerholz D.K., Sutterwala F.S., Narumiya S., Perlman S. Virus-induced inflammasome activation is suppressed by prostaglandin D2/DP1 signaling. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E5444–E5453. doi: 10.1073/pnas.1704099114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilo, J. (2002) Pattern discovery from biosequences.
- Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A., Peters B. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(Database issue):D405–12. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin T., Rouet-Benzineb P., Reuter N., Laburthe M. Orexins and their receptors: structural aspects and role in peripheral tissues. Cell. Mol. Life Sci. 2003;60:72–87. doi: 10.1007/s000180300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Liu I.J., Lu R.M., Wu H.C. Advancement and applications of peptide phage display technology in biomedical science. J. Biomed. Sci. 2016;23:8. doi: 10.1186/s12929-016-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.J., Kula T., Xu Q., Li M.Z., Vernon S.D., Ndung'u T., Ruxrungtham K., Sanchez J., Brander C., Chung R.T., O'connor K.C., Walker B., Larman H.B., Elledge S.J. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandian A., Forsstrom B., Haggmark-Manberg A., Schwenk J.M., Uhlen M., Nilsson P., Ayoglu B. Whole-proteome peptide microarrays for profiling autoantibody repertoires within multiple sclerosis and narcolepsy. J. Proteome Res. 2017;16:1300–1314. doi: 10.1021/acs.jproteome.6b00916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material