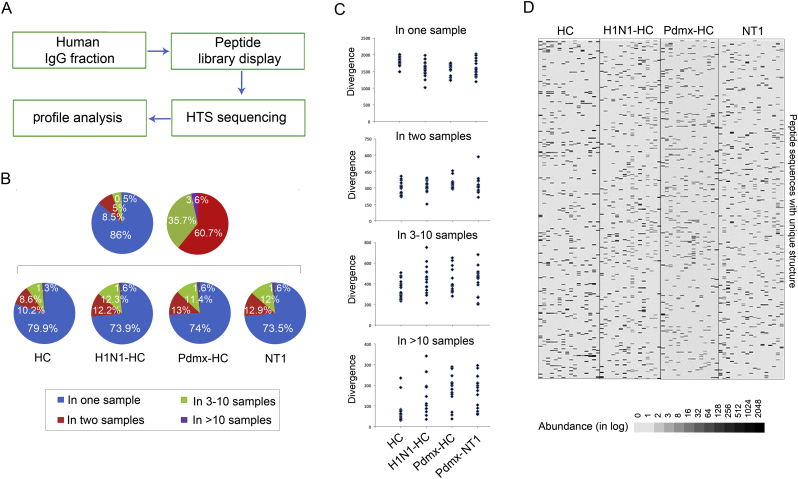
Fig. 1.
Humoral immune response studied using the mimotope-variation analysis (MVA) method. A. Schematic drawing of the workflow in MVA. MVA is a high-throughput random peptide phage display analysis. A random peptide display library (PhD12) was used which contained 10^9 different 12-mer peptide sequences introduced to the N-terminus of the phage major coat protein pIII (NEB). For MVA, sample-specific IgG proteins (antibodies, Human IgG fraction) present in human sera of interest are allowed to interact with the phage-displayed peptides and the IgG-phage complexes were captured to protein G magnetic beads, while the unbound phages were washed away (Peptide library display). Captured phages were lysed and DNA amplified with primer sequences containing a tag with a unique barcode sequence and the final amplicons were pooled for NGS analysis (HTS sequencing). The primer set homologous to the M13KE vector sequences that flank the random peptide coding sequence was used to amplify a 50-bp fragment. Data analysis to classify peptides that were specific to Pmdx-infected, -vaccinated and NT1-diseased individuals was carried out by comparing the profiles of peptides (mimotopes) from diseased to those from non-diseased (Peptide profile analysis). On average, MVA generated 1.8 million peptide sequences with unique structure (divergence) totaling 2.8 million peptide sequences in abundance (total abundance; number of reads) per sample. Altogether, a peptide data set with >16 million sequences (Totpep) with unique structure was generated.
B. Analysis of peptides revealed highly divergent patterns (immunoprofiles) across study cohorts. The fraction of top 2500 peptides with unique structure and highest values of abundance - reflecting the peaking immune reactivity of each sample - was analyzed for variance. Top2500 peptide dataset contained altogether 160,000 sequences out of which 121,142 were unique. Pie charts display the sequence distribution of unique peptides across all samples analyzed. The left pie (blue) displays the proportion of shared vs. unique peptides: ~86% were unique to one individual whereas ~14% of the peptide sequences were shared between samples, out of these ~8.5% were common to 2 samples, 5% to 3–10 samples and 0.5% were detected in >10 samples. The right pie (red) displays the distribution of shared 16,844 peptide sequences out of which ~60.7% were common to 2 samples, 35.7% to 3–10 samples and 3.6% were seen in >10 samples. The four pie charts (below) exemplify the peptide profile structures in different clinical cohorts. The size of each pie piece is proportional to the number of unique peptides common to one or more samples of a clinical cohort. Blue - represents unique peptides, red - the most shared.
C. Individual variation in peptide divergence is characteristic to all immunoprofiles. Top 2500 peptides were analyzed to assess the range of individual peptide variation across study cohorts. Blue dots mark peptide divergence in a single sample. As indicated, between one to two thousand peptides were individual-specific, whilst the most common peptides (shared by >10 individuals) ranged in divergence from tens to 350 across samples. Range of unique peptide variations was similar across all study samples.
D. Heat map image of a random fragment of MVA profile encompassing 400 peptides across study samples. Peptide profiles were individual-specific with a highly varying abundance. Each column represents the peptide profile of a single individual, and each line represents a peptide with a unique primary structure. Abundance is presented as counts in logarithmic scale (in log); black colour depicts peptides captured at higher abundance, and white those at lower abundance. Shown are peptide profiles that were common to 3–10 individuals. Abbreviations: Abundance – peptide frequency; Divergence – all unique peptides; HC- healthy control; H1N1-HC – H1N1 infected; Pdmx-HC- Pandemrix-vaccinated; NT1- narcolepsy type 1 (including 10 Pandemrix-induced NT1 samples).