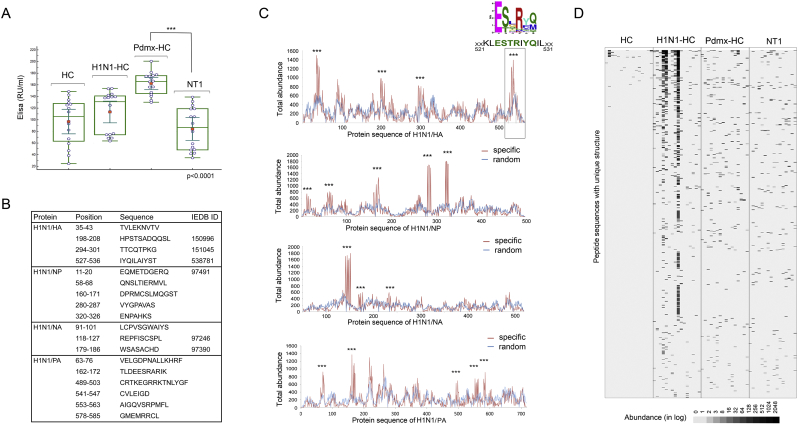
Fig. 2.
A novel common epitope of HA antigen of A/H1N1 in seasonal infection carriers and Pdmx-vaccinated individuals, encompassing a proven T cell- antigenic region.
A. The humoral response to seasonal flu (A/H1N1 and A/H3N2) was relatively weaker in NT1-diseased as compared with Pdmx-HC individuals as determined by using a commercial ELISA test (p < .001). The presence of IgG antibodies against HA and H3N2 was assessed in sera samples of HC, H1N1-HC, Pdmx-HC and Pdmx-NT1 by ELISA (Influenza virus type A IgG ELISA, Euroimmun). The HC samples were collected prior to A/H1N1 outburst in Estonia, before the fall 2009. Blue circles mark individuals of the study cohorts; red dots mark the mean values; lines depict median values; inner whiskers mark confidence interval for the mean; boxes mark upper and lower quartiles; outer whiskers mark the maximum and minimum values (excluding the outliers). P-values were calculated by ANOVA and are marked with asterisks. The cut-off value for ELISA was 16 RU/mL. Labels at the top of box plots demark the clinical origin of the sample.
B. MVA predicted H1N1 epitopes partially overlapped with previously described H1N1 (A/California/08/2009(H1N1)) B cell specific epitopes from IEDB (http://www.iedb.org/). Top2500 peptide dataset containing 121,142 unique sequences was used to delineate the predominant epitopes of H1N1/HA (GI: 238,623,304), H1N1/NA (NA, GI:758899360), H1N1/NP (NP, GI:229891180) and H1N1/PA proteins in study samples. 9657 peptides from the studied dataset satisfied the selection criteria that these were not present in the HC samples. Specific alignment profiles for each of the A/H1N1 protein antigens were calculated with the criterion that the abundance of a peptide was to be 2-fold higher over random.
C. MVA immunoprofiles predicted a novel epitope in the C-terminal region of HA encompassing amino acids 521–531 and with the sequence ESxRxQ that was common to both seasonal infection carriers and Pdmx-vaccinated individuals. The graphs show antigen-specific profiles of overall peptide abundance where the number of peptides were counted for each amino acid position for the following proteins: hemagglutinin (H1N1/HA, C4RUW8), neuraminidase (H1N1/NA, C3W6G3), nucleoprotein (H1N1/NP, B4URE0) and polymerase acidic protein (H1N1/PA_I6THC5). Amino acid sequence of the proteins is depicted on the x-axis. Marked with asterisks are regions where set calculation criteria were satisfied. Detailed analysis of immunoprofiles of H1N1 antigens revealed a novel immunogenic region of HA encompassing amino acids 521–531 that corresponds to the earlier experimentally determined A/Puerto Rico/8/1934(H1N1) HA520–530 CTL epitope (Gianfrani et al., 2000) and is partially overlapping with broadly reactive CD4+ T cell epitope: HA527–541 of A/California/04/2009(H1N1) (Schanen et al., 2011). Peptides aligning to 521–531 of HA cluster to a minimal consensus sequence E[ST].R.[QM] by sequence homology alignment.
D. Heat map image of immunoprofiles of peptides with consensus E[ST].R.[QM] across study samples. The total peptide data set was examined for the peptides with unique structure clustering to E[ST].R.[QM] motif. About 700 peptides with enriched abundance in H1N1-HC, Pdmx-HC or Pdmx-NT1 samples were found to cluster to the motif. The data of 700 peptides is presented on the heat map image. Each line represents peptides with unique sequence structure. The colour intensity of each cell corresponds to the peptide abundance (presented in log value). Black represents peptides captured at higher abundance whereas white represents peptides captured at lower abundance. Each column represents a peptide profile from a single sample. Labels at the top of the panels indicate the clinical origin of the sample. Abbreviations: Random alignment – amino acid sequences of peptides under analysis were randomized and aligned to respective protein coding sequence; Total abundance - the number of peptides counted for defined amino acid positions; HC– healthy control; H1N1-HC – H1N1 infected; Pdmx-HC- Pandemrix-vaccinated; NT1- narcolepsy type 1 (including 10 Pandemrix-induced NT1 samples).