ABSTRACT
Four of eleven affected dogs died despite aggressive treatment during a 2015 focal outbreak of hemorrhagic gastroenteritis following a stay in a pet housing facility. Routine diagnostic investigations failed to identify a specific cause. Virus isolation from fresh necropsy tissues yielded a calicivirus with sequence homology to a vesivirus within the group colloquially known as the vesivirus 2117 strains that were originally identified as contaminants in CHO cell bioreactors. In situ hybridization and reverse transcription-PCR assays of tissues from the four deceased dogs confirmed the presence of canine vesivirus (CaVV) nucleic acids that localized to endothelial cells of arterial and capillary blood vessels. CaVV nucleic acid corresponded to areas of necrosis and hemorrhage primarily in the intestinal tract, but also in the brain of one dog with nonsuppurative meningoencephalitis. This is the first report of an atypical disease association with a putative hypervirulent vesivirus strain in dogs, as all other known strains of CaVV appear to cause nonclinical infections or relatively mild disease. After identification of the CU-296 vesivirus strain from this outbreak, four additional CaVV strains were amplified from unrelated fecal specimens and archived stocks provided by other laboratories. Broader questions include the origins, reservoir(s), and potential for reemergence and spread of these related CaVVs.
KEYWORDS: calicivirus, canine, vesivirus
INTRODUCTION
Vesiviruses constitute a genus in the Caliciviridae family. Other genera within the family include Lagovirus, Nebovirus, Norovirus, and Sapovirus. The type species of the vesivirus genus is the now eradicated vesicular exanthema of swine virus that emerged in the United States in the 1930s and was later attributed to a likely cross-species transmission of San Miguel sea lion virus (1). The ability of the marine-origin vesiviruses to cross species boundaries and establish infections in diverse hosts is a hallmark of the genus (2).
One of the most well-studied vesiviruses is feline calicivirus (FCV), a common pathogen in domestic cats that displays a spectrum of clinical signs but primarily upper respiratory tract infections. Most are subclinical or mild, but infrequently a virulent systemic form has been reported (vsFCV) (3). Variants of FCV have been identified in dogs associated with gastroenteritis and glossitis (4–9). Schaffer et al. (10) isolated a novel calicivirus from a dog in Tennessee with gastroenteritis and neurologic signs and found that it was serologically distinct from FCV and other vesiviruses. Thereafter, two caliciviruses were isolated from canine genital lesions (11). These viruses were not cross-neutralized by antiserum specific to the virus isolated by Schaffer et al. (10) nor to FCV. A calicivirus (strain 48) was isolated from a dog with diarrhea in Japan (12, 13). Cross-neutralization experiments showed that it was distinct from three strains of FCV (12). Experimental infections with the Tennessee and strain 48 isolates failed to reproduce disease (10, 12).
In 2003 a vesivirus was identified as a contaminant in CHO cell cultures in a pharmaceutical facility in Germany (14). Two additional strains, Geel and Allston, were subsequently discovered in production facilities in Belgium and the United States (15). The CHO cell isolates are all closely related and often collectively referred to as vesivirus strain 2117. This virus is a continuing concern for the biopharmaceutical industry, and several commercial PCR kits are marketed for routine screening of cell cultures. The source of the contamination was never identified, but bovine serum products were suggested as a likely source (14). More recently, metagenomic analysis of a canine gastroenteritis case identified a partial sequence from a 2117-like virus (16). Martella et al. (17) screened canine fecal samples with broadly reactive PCR primers and identified 2117-like viruses in 65% of shelter-housed animals, but in only 1.1 and 3.5% of gastroenteritis cases and otherwise healthy household dogs, respectively.
In this study, four of eleven dogs with hemorrhagic diarrhea and vomiting died within 24 to 72 h of onset of clinical signs despite immediate and aggressive therapy. A 2117-like vesivirus (CU-296) common to the deceased dogs was identified and in situ hybridization (ISH) revealed a systemic infection with the viral RNA primarily localized to endothelial cells in conjunction with hemorrhage. The clinical presentation and similarities with outbreaks of vsFCV are described.
MATERIALS AND METHODS
Diagnostic investigation.
All but one affected dog were treated at the Aldie Veterinary Hospital (South Riding, VA). The other (dog 1) was treated at another emergency care facility (The Life Center, Leesburg, VA) since it developed clinical signs after returning home. Initial diagnostic testing performed on site included complete blood counts, serum chemistry, prothrombin time and activated partial thromboplastin tests, fecal smears/floats, cytology, radiography, and ultrasound FAST scans. Canine circovirus (CCV) testing was performed by a commercial diagnostic service (IDEXX Laboratories, Westbrook, ME). Minerals analysis, general toxicology, and immunofluorescence assays (IFA) for CCV, canine adenovirus (CAdV) and canine enteric coronavirus (CCoV) were performed by a referral diagnostic laboratory (Michigan State University, Diagnostic Center for Population and Animal Health, Lansing, MI). Specimens from dogs that were treated and released were not available and thus were not included in this study. Dogs 1, 2, and 3 were necropsied at Aldie Veterinary Hospital. Dog 4 was sent to the Virginia State Warrenton Regional Animal Health Laboratory (WRAHL; Warrenton, VA). Selected tissue samples from dogs 1 to 3 were obtained at necropsy, fixed in 10% neutral buffered formalin, and submitted for routine histopathology examination (Antech Diagnostics, Lake Success, NY). The following formalin-fixed and paraffin-embedded (FFPE) tissue samples were examined: liver, kidney, stomach, duodenum, jejunum, ileum, and colon from dog 1; small intestine and lymph node from dog 2; and heart, lung, liver, spleen, kidney, stomach, duodenum, jejunum, ileum, cecum, colon, and pancreas from dog 3. Gastrointestinal (GI) PCR panels, including Campylobacter coli, Campylobacter jejuni, CCoV, canine parvovirus (CPV), Clostridium difficile toxins A/B, Clostridium perfringens enterotoxin, Cryptosporidium spp., Giardia spp., and Salmonella spp., were performed on intestinal samples collected from dogs 1 to 3 by a commercial diagnostic service (Antech Diagnostics). The body of dog 4 was submitted to WRAHL for complete necropsy and diagnostic evaluation, including histopathology, bacteriology, and IFA for CAdV, CCoV, and CPV. For histopathology of dog 4, FFPE tissue sections of the brain, heart, trachea, lung, liver, spleen, kidney, urinary bladder, tongue, stomach, small and large intestines, pancreas, lymph nodes, eyelids, thyroid gland, adrenal gland, and bone marrow were examined. Subsets of FFPE tissue samples taken from dogs 1 to 4 were forwarded from the respective diagnostic laboratories to the New York Animal Health Diagnostic Center (AHDC) for further assessment by a board-certified veterinary pathologist (G.E.D.) using routine histopathology, immunohistochemical (IHC) staining, and a novel and highly sensitive CaVV-specific ISH assay. Virus isolation and molecular analysis of fresh samples of the brain, lung, liver, small intestine, and colon obtained from dog 4 were performed at the AHDC. IFA for FCV was performed using monoclonal antibody S1-9 (Custom Monoclonal Antibodies International, West Sacramento, CA).
Virus isolation and cell culture.
Suspensions containing approximately 10% lung, intestine or brain from dog 4 were prepared in Eagle minimum essential medium (MEM-E) with 0.5% bovine serum albumin, and antibiotic mix (200 U/ml penicillin, 200 μg/ml streptomycin, 2.5 μg/ml amphotericin B, 10 μg/ml ciprofloxacin). Tissues were homogenized then centrifuged at 1,000 × g for 15 min. The intestinal homogenate was additionally passed through 1.2-μm-, 0.8-μm-, and 0.45-μm-pore size filters. Aliquots (1 ml) of lung and intestine homogenates were overlaid on semiconfluent monolayers of an immortalized canine kidney cell line, CK-DE2 (AHDC, Cornell University). The homogenate was allowed to remain on the monolayer for 1 to 3 h and then rinsed twice with phosphate-buffered saline, followed by incubation at 37°C in a MEM-E, 5% FBS, and antibiotic mix. Cells were subcultured using trypsin at 5- to 7-day intervals, and new monolayers were established at a 1:6 split ratio. Subcultures were observed daily for the presence of cytopathic effect. Virus was also cultured on an immortalized canine skin cell line (AHDC, Cornell University) in MEM-E, 5% FBS, 200 U/ml penicillin, and 200 μg/ml streptomycin.
TEM.
One 75-cm2 flask of infected CK-DE2 cells was frozen at −80°C and thawed at 37°C. The culture was centrifuged at 2,000 × g for 10 min at 4°C to remove cellular debris. Virus was pelleted at 71,000 × g for 2 h at 4°C and resuspended in 1.0 ml of phosphate-buffered saline for negative-staining transmission electron microscopy (TEM; Texas Veterinary Medical Diagnostic Laboratory [TVMDL], College Station, TX).
Real-time PCR assays.
For detection of CU-296 and related vesivirus strains in clinical samples, three real-time reverse transcription-PCR (RT-PCR) hydrolysis probe assays were designed using Primer Express (Applied Biosystems, Foster City, CA). Primer and probe sequences were determined based on consensus of the alignment of available 2117 strains and CU-296 (see Table S1 in the supplemental material). Three primer sets were used: 3461F/3553R and 3443F/3553R targeting open reading frame 1 (ORF1) and 5750F/5823R beginning in the 3′ end of ORF1 and spanning the ORF1/ORF2 junction. Nucleic acids from unfixed specimens were extracted using a QIAamp viral RNA kit (Qiagen, Valencia, CA). For FFPE specimens, several sections totaling 50 μm in thickness were extracted using a RecoverAll total nucleic acid isolation kit (Ambion, Foster City, CA). Separate exogenous control reactions (18) were run after the addition of a MS2 phage standard to the extraction buffer used with all specimens. Single-tube reactions used a PATH ID multiplex one-step RT-PCR kit (Applied Biosystems) StepOnePlus system (Applied Biosystems) with cycling conditions of 48°C for 10 min, followed by 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 60 s. Then, 4 μl of purified nucleic acid was used in reaction mixtures containing 400 nM concentrations of each primer and a 120 nM concentration of probe.
Conventional RT-PCR and sequencing.
For initial identification, a passage 4 isolate of CU-296 cultured on CK-DE2 cells was extracted using a Qiagen QIAamp viral RNA extraction kit. The RNA was amplified using a SuperScript III One Step RT-PCR kit (Life Technologies, Carlsbad, CA) with RNA polymerase-specific primer sets 493F/526R and 290F/289R (see Table S2 in the supplemental material). Whole-genome sequencing was initiated upon confirmation that the isolate was a vesivirus. First-strand cDNA was synthesized using SuperScript IV reverse transcriptase (Life Technologies) with overlapping primer sets for amplification and sequencing designed using alignments of the previously published 2117 virus sequences and newly generated CU-296 sequence (see Table S2 in the supplemental material). Amplicons were generated with a Q5 Hot Start High Fidelity 2× master mix (New England BioLabs, Ipswich, MA) using reaction conditions of 40 cycles of 98°C for 10 s, a 2° increment gradient of 40 to 50°C for 30 s, and extension at 72°C for 90 s on a Veriti thermocycler (Applied Biosystems). Individual reactions were run at each gradient temperature. Genome termini of CU-296 were determined using a RACE system kit (Life Technologies). Full-length ORF2 capsid genes were amplified and sequenced from archival strain 143 (courtesy of Pamela J. Ferro, TVMDL), two field strains amplified from fecal samples (CU-47 and CU-103), and isolate LSU 96-6931 (Louisiana State University) (Table 1). Specific primers used for VP2 sequencing are listed in Table S2 in the supplemental material. All sequencing was performed using the Sanger method (Biotechnology Resource Facility, Cornell University).
TABLE 1.
Summary of CaVV strains analyzed in this studya
| Strain | Yr | Location | Tissue | Region sequenced |
|---|---|---|---|---|
| LSU-96-6931 | 1996 | LA | Brain | Capsid gene |
| 143 | 1997 | NA | NA | Capsid gene |
| CU-296 | 2015 | DC | Intestine | Genome, complete |
| CU-47 | 2016 | NY | Feces | Capsid gene |
| CU-103 | 2017 | NY | Feces | Capsid gene |
NA, not available.
Sequence analysis.
Sequence construction and analysis were performed using the Lasergene Molecular Biology Suite (DNASTAR, Madison, WI). Sequence similarities were identified using BLAST analysis at GenBank, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/nucleotide). Evolutionary analyses were conducted in MEGA7 (19). Sequences were aligned with ClustalW (20), phylogeny was analyzed using the neighbor-joining method (21), and confidence limits of trees were calculated using bootstrap analysis (22). Evolutionary distances were calculated using the maximum-likelihood method (23). Amino acid distance analysis was visualized using SSE v1.2 (24).
Histopathology and IHC.
Selected FFPE tissues, including duodenum from dog 1, small intestine and lymph node from dog 2, lung from dog 3, and all tissues from dog 4 were sectioned at a 5-μm thickness and stained with hematoxylin and eosin (H&E). The presence of immune and inflammatory cells in sections of the brain from dog 4 was assessed by IHC analysis. Briefly, sections of tissue were processed for heat epitope retrieval (Leica Bond Max staining system; Leica Biosystems) and incubated with the respective primary antibodies for 10 to 30 min at room temperature or the corresponding mouse IgG or rabbit IgG (Invitrogen, Carlsbad, CA) as a negative control. An automated IHC stainer (Leica Bond-Max; Leica Biosystems) and commercial polymeric horseradish peroxidase (Bond Polymer Refine Detection; Leica Biosystems) or alkaline phosphatase (Leica Bond Polymer Refine Red Detection; Leica Biosystems) linker antibody conjugate detection system and hematoxylin counterstain were used. Sections of normal lymphoid tissue served as positive control for immunolabeling of T and B lymphocytes, dendritic cells, and macrophages. The following primary antibodies to cellular markers were used: plasma cells and B lymphocytes were labeled with a rabbit polyclonal IgG anti-human CD20 (PA5-16701; Thermo Fisher Scientific, Rockford, IL) and a mouse monoclonal IgG1, Pax-5 (1EW) directed at a transcription factor for B-lymphocyte development (PA0552; Leica Biosystems Newcastle, Ltd.); T lymphocytes were labeled with the mouse monoclonal IgG2a, clone LN10 (PA0553; Leica Biosystems Newcastle, Ltd.), anti-human T cell antigen receptor complex CD3; tissue histiocytes, circulating and emigrated neutrophils, monocytes, and a subset of reactive tissue macrophages were labeled with the mouse monoclonal IgG1, clone MAC387 (M0747; DakoCytomation), anti-human myeloid/histiocyte antigen; activated macrophages and dendritic cells (25) were labeled with a rabbit polyclonal antibody raised against a conserved human, rat and mouse synthetic peptide of the ionized calcium binding adaptor molecule 1 (Iba-1; 019-19741; Wako Chemicals GmbH, Neuss, Germany); and tissue histiocytes expressing macrophage scavenger receptor-A (CD204) were labeled with a mouse monoclonal IgG1, clone SRA-E5 raised against a recombinant protein of human type 1 MSR-A (KAL-KT022; Cosmo Bio Co., Ltd.).
ISH.
A highly sensitive ISH method (26) (RNAscope; Advanced Cell Diagnostics [ACD], Hayward, CA) was used for cellular localization of CaVV in FFPE tissue sections. For this study, a novel 20 Z pair probe (ACD, catalog no. 472281) targeting three conserved regions of the genome, as determined by multiple sequence alignments of 2117-like strains (see Table S3 in the supplemental material), was designed so as to prevent nonspecific hybridization to canine genomic sequences. A potential to hybridize to FCV and related vesiviruses is recognized since there are regions of 67 to 74% similarity over approximately 40% of the sequence submitted for probe design. However, there are few contiguous regions of 20 bp in length (the approximate span required for 1 Z probe pair), and the exact sequences of the Z probe pairs are held proprietarily, so in silico assessment of cross-reactivity is not practical. If used diagnostically, cross-reactivity to other vesiviruses would need to be determined empirically, although nonspecific signal would likely be weak. The use of control uninfected CK-DE2 cells and cells infected with CU-296 confirmed the specificity of the ISH probe as used for this study. For ISH, 5-μm-thick sections of FFPE tissue were placed on charged slides and processed using an RNAscope 2.5 HD Brown assay according to the manufacturer's protocol (27). After deparaffinization, tissue sections were permeabilized and hybridized with the CaVV probe, followed by signal amplification, binding of a chromogenic agent, and counterstaining with hematoxylin, allowing direct visualization of CaVV-infected cells by light microscopy. Positive staining was identified as brown, punctate areas within the nuclei and cytoplasm of infected cells. Negative- and positive-control ISH probes included a bacterial gene Dap-B and ubiquitin C (CI-UBC), respectively (ACD).
Accession number(s).
The full isolate name is CU/296/15/USA, and it is used in abbreviated form as CU-296. The CU-296 genome is entered in GenBank under accession number MF978270. The capsid gene sequences of strains 143, CU-47, CU-103, and LSU 96-6931 are entered in GenBank under accession numbers MG213713 to MG213716. The accession numbers of related vesiviruses used in the analysis were as follows: Allston/08 and Allston/09 (GQ475301, GQ475302), Bari/212/07/ITA (JN204722), Geel/08 (GQ475303), strain 2117 (AY343325), and strain 48 (AB070225).
RESULTS
Clinical findings.
In August 2015, 11 dogs at a kennel in the Washington, DC, metropolitan area presented with acute signs of hemorrhagic gastroenteritis. All dogs were between 1 and 5 years of age, up-to-date with common vaccines, and otherwise healthy prior to the onset of clinical signs. Seven dogs were sufficiently stable to be released following routine outpatient medical management or less than 48 h of hospitalization. Four dogs rapidly decompensated over the course of 24 to 72 h and died (n = 1) or were euthanized due to poor prognosis (n = 3). The clinical signs of the severely affected dogs are summarized in Table 2.
TABLE 2.
Clinical history of four fatal cases associated with CaVV CU-296 infection
| Case | Gendera | Breed | Age (yrs) | Onset date | Clinical presentation |
|---|---|---|---|---|---|
| 1 | NM | Beagle mix | 3 | 8/4/2015 | Severe acute vomiting, bloody diarrhea, cardiopulmonary arrest within 6 h of presentation; euthanized 8/5/2015. |
| 2 | NM | Golden doodle | 1 | 8/2/2015 | Acute vomiting, bloody diarrhea, severe profuse hematochezia, depression, obtunded, nystagmus and seizures, cardiopulmonary arrest; resuscitation unsuccessful 8/4/2015. |
| 3 | SF | Labrador retriever | 2 | 8/20/2015 | Acute vomiting, diarrhea, hematochezia, severe lethargy; decompensated very quickly, obtunded mentation, arrhythmia; euthanized 8/21/2015. |
| 4 | NM | Cockapoo | 1 | 8/24/2015 | Acute bloody diarrhea, hematochezia, gastric reflux, bleeding complications (suspected disseminated intravascular coagulation), progressive depression; euthanized 8/26/2015. |
NM, neutered male; SF, spayed female.
Laboratory findings.
Diagnostic specimens from five of the eleven affected dogs were submitted for a GI PCR panel with results of three negative (two deceased, one survivor), one Giardia positive (survivor), one CCoV positive (survivor). Additional preliminary results included three dogs positive (one deceased, two survivors) by fecal culture for coliforms, Yersinia enterocolitica, and Campylobacter jejuni; four dogs negative for CCV (two deceased, two survivors); one canine distemper virus PCR-negative dog (dog 2); one dog negative for leptospirosis (survivor); and one dog that tested normal for pancreatic lipase (survivor). Seven of the dogs tested were negative for intestinal parasites with an in-house fecal exam. All dogs that were tested (eight of eleven) had relatively normal blood counts aside from various degrees of hemoconcentration. All chemistry panels (eight of eleven) were relatively within normal limits, aside from two dogs with a mildly elevated total bilirubin level and one survivor with an elevated gamma-glutamyl transferase level (25 U/liter). Three of the four dogs that died had partial thromboplastin and prothrombin time testing on presentation; two of the three were within normal range, and the fourth was mildly elevated. Dog 4 was negative for Salmonella by bacterial culture and negative for CAdV, CCoV, and CPV by IFA. There were no significant findings by aerobic and anaerobic bacteriological cultures of the lung, liver, lymph node, or intestine from dog 4. Toxins from food sources were considered, but ultimately not supported. Although several of the 11 affected dogs received one of three varieties of feed from the same manufacturer, the vast majority of the dogs receiving the same feed showed no clinical signs. Although there were positive test results for some dogs, no two dogs with the same clinical signs tested positive for the same disease, or the clinical signs of the more severely affected dogs did not match the diagnosis of the lesser affected dogs. Only tissues from the four deceased dogs were available for further testing. Diagnostic specimens from the survivors were not retained by the testing labs. Postrecovery serum samples from the survivors were not available.
Histopathology and IHC findings.
The initial histopathology assessment of tissues obtained during field necropsy of dogs 1 to 3 revealed lesions restricted to the intestinal tract. The initial diagnosis for dog 1 was idiopathic hemorrhagic enteritis, while dogs 2 and 3 had lesions suggestive of diet-induced chronic enteropathy and small intestinal congestion and hemorrhage, respectively. Further assessment of selected FFPE tissues confirmed small-intestinal lesions in dogs 1 and 2; however, the changes were interpreted to be severe and consistent with acute mucosal necrosis and hemorrhage (Fig. 1A). On closer examination, areas of mucosal necrosis with frequent fibrinoid necrosis of arterial blood vessel walls (Fig. 1B and C), along with hyaline thrombi within villous lamina propria capillaries were noted. A section of lymph node from dog 2 also showed acute sinus hemorrhage with erythrophagocytosis and edema. The only tissue from dog 3 that was available for further examination was a section of lung that showed mild, acute, multifocal interstitial perivascular and airway lumen hemorrhage. In contrast, dog 4 had an extensive set of tissues that revealed severe lesions primarily within the gastrointestinal tract and brain. The changes in the small intestine were similar to those seen in dogs 1 and 2; however, these changes were more extensive and severe in the duodenum, with involvement of all segments of the small and large intestines. There were also severe lymphoid depletion of Peyer's patches in the ileum and multifocal acute hemorrhage within the tunica muscularis throughout the small and large intestines. Multifocal acute hemorrhage also was present in the heart, the cortex of the adrenal gland, and mucosa of the urinary bladder. The lung of dog 4 had a locally extensive aspiration pneumonia and a section of lymph node had acute sinus hemorrhage similar to that seen in dog 2. Sections of cerebrum and cerebellum from dog 4 showed frequent perivascular cuffs composed of mostly lymphocytes and macrophages within the gray and white matter but also within the meninges and choroid plexus (Fig. 2E). The IHC staining confirmed that perivascular cuffs in the brain consisted of mixed mononuclear cell infiltrates composed of T lymphocytes with membrane and cytoplasmic CD3 immunoreactivity admixed with macrophages with membrane and cytoplasmic Iba-1 and CD204 immunoreactivity. Staining of brain sections for the presence of CD20, Pax-5, and MAC387 showed no immunoreactivity for these antigens. Sections of brain from dog 4 also showed extensive capillary endothelial swelling with prominent fibrinoid necrosis of arterioles, mild perivascular hemorrhage, and scattered glial nodules within the gray matter of the cerebrum. The glial nodules showed immunoreactivity for the presence of Iba-1 antigen. On the basis of these findings, a viral etiology was suspected, and this prompted further diagnostic investigation.
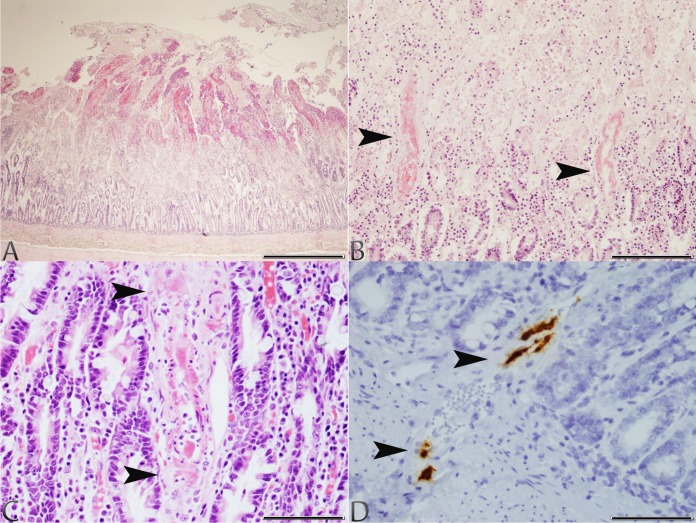
FIG 1.
Photomicrographs of tissues taken from dogs 1 and 2 with typical lesions of CaVV infection. (A) Small intestine, dog 1: severe, acute superficial mucosal necrosis and hemorrhage (H&E stain; scale bar, 850 μm). (B) Small intestine, dog 1: fibrinoid necrosis of the arterial blood vessel walls (arrowheads) at the junction of an area of acute superficial mucosal necrosis and hemorrhage (H&E stain; scale bar, 170 μm). (C) Small intestine, dog 2: fibrinoid necrosis of an arterial blood vessel wall (arrowheads) within the lamina propria (H&E stain; scale bar, 85 μm). (D) Small intestine, dog 2: CaVV nucleic acid signal in endothelial cells of arterial blood vessels (arrowheads) at the junction between the lamina propria and submucosa (ISH; scale bar, 85 μm).
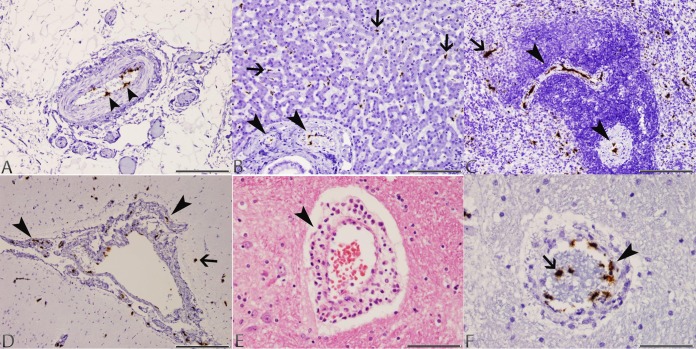
FIG 2.
Photomicrographs of tissues taken from dog 4 with typical lesions of CaVV infection. (A) Mesentery: CaVV nucleic acid signal in endothelial cells of a medium-sized arterial blood vessel (arrowheads) in the mesenteric attachment of the ileum (ISH; scale bar, 170 μm). (B) Liver: CaVV nucleic acid signal in endothelial cells of portal arterial blood vessels (arrowheads) and Kupffer cells along sinusoids (arrows) (ISH; scale bar, 170 μm). (C) Spleen: CaVV nucleic acid signal in endothelial cells of arterial blood vessels within the white pulp (arrowheads), and mononuclear cells within the red pulp (arrow; ISH; scale bar, 170 μm). (D) Brain: CaVV nucleic acid signal in endothelial cells of meningeal arterial blood vessels (arrowheads), and scattered glial cells in the cortex (arrow) (ISH; scale bar, 330 μm). (E) Brain: perivascular cuff of lymphocytes and macrophages within the cerebrum (arrow) (H&E; scale bar, 85 μm). (F) Brain: CaVV nucleic acid signal in endothelial cells (arrowhead) and circulating intravascular leukocytes (arrow) in a medium-sized arterial blood vessel (ISH; scale bar, 85 μm).
Virus culture and identification.
Cultures of CK-DE2 cells inoculated with tissue homogenate from dog 4 showed complete disruption of the cell monolayer within 3 days postinoculation. Passage of the supernatant to fresh cultures produced an overnight cytopathic effect with rounding and detachment. The virus was also cultured on an immortalized canine skin cell line. Canine A-72 fibroblasts, Madin-Darby canine kidney (MDCK) cells, and Vero cells did not support replication. TEM identified a calicivirus in pelleted cell lysates. An IFA for FCV was negative. PCR using primers designed for strain 48 (CaCV48-493F/526R) (17) and a set more broadly specific to caliciviruses (CV-290F/CV-289R) (28) (see Table S2 in the supplemental material) yielded overlapping amplicons. A BLAST search of the sequences identified a 2117-like vesivirus. The same primer sets generated amplicons from strain 143 and LSU 96-6931. Additional screening for CaVV by RT-PCR was conducted on 144 fecal specimens of convenience from dogs from eight states in the United States (NY, PA, DE, CT, MA, SC, VT, and CA). Submissions were categorized as GI disease cases (21.5%), other illness (13.9%), routine screening (16.7%), or without clinical history and health status unknown (47.9%). Two new strains were identified and designated CU-47 and CU-103. Both were from dogs of undetermined health status. Attempts to propagate lyophilized strain 143 and fecal strains CU-47 and CU-103 in CK-DE2 cells were unsuccessful. LSU/96-6931 was successfully cultured on CK-DE2 cells.
Genome analysis.
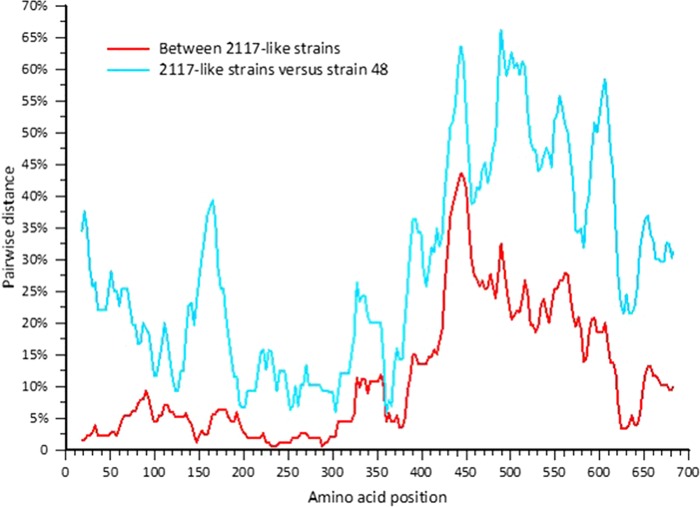
Phylogenetic analyses of CU-296 showed it was closely related to the CHO cell 2117 strains, including Bari/212/07/ITA (Fig. 3). The genome is 8,447 nucleotides in length, with an overall identity of 89 to 90% to the 2117 strains but only 72.6% to strain 48. Three ORFs were confirmed to encode a polyprotein product of 1,931 amino acids (aa), a capsid precursor protein (VP1) of 690 aa, and a minor structural protein (VP2) of 134 aa. Amino acid motifs common between caliciviruses and vesivirus 2117 are conserved in CU-296 (14). Differences that might speculatively point to an alteration of phenotype were not identified in any of the translation products. Compared to the CHO cell 2117 strains, the translation products of CU-296 and Bari/212/07/ITA showed a high degree of conservation for ORFs 1 and 3, with 98 to 99% identity in all cases (Table 3). The ORF1 polyprotein sequence was compared to the only other full-length canine-origin 2117-like sequence, Bari/212/07/ITA, and showed 13 aa differences, 10 being scored as conserved or very highly conserved. Two of the nonconserved residues are located in the most variable region of the ORF1 polyprotein located at aa 155 to 173 in the carboxy-terminal region of the nonstructural (NS) protein. One amino acid difference, A→V at aa 99, is the only amino acid in the ORF1 polyprotein that is unique to CU-296 compared to the group of 2117-like viruses. The VP2 minor structural protein has one unique amino acid difference with the 2117-like viruses: V→A at aa 3. High variation in the VP1 capsid is a hallmark of vesiviruses with highest diversity in the carboxy-terminal third corresponding to the vesivirus E and F capsid regions (29, 30) (Fig. 4). Mean pairwise distances between the 2117-like virus group and strain 48 also indicate significant variation in the capsid core region B (Fig. 4). VP1 of CU-296 has the highest amino acid identity with the LSU 96-6931 and prototype 2117 strains and the lowest identity among the 2117-like strains to CU-103 (Table 4). Other than the two Allston strains that are presumed to be of the same origin, none of the pairwise identities are higher than between CU-296 and LSU 96-6931 (data not shown). Phylogenetic relationships between the capsid proteins of canine origin CaVV and related 2117 strains are depicted (Fig. 5).
FIG 3.
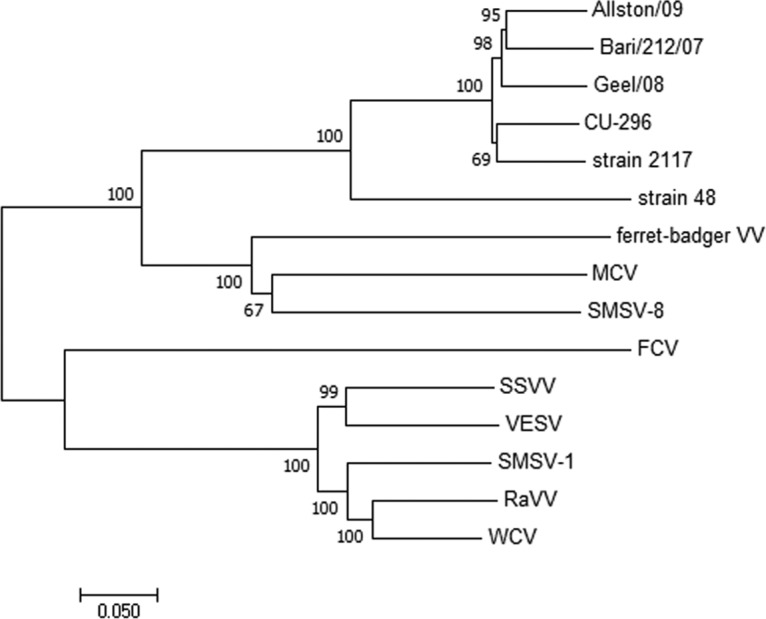
Relationship of full-length vesivirus genomes based on the neighbor-joining method (21). Percentages of 500 replicate trees in which the sequences clustered together are indicated at the nodes (22). Horizontal branch lengths are drawn to scale of the number of nucleotide substitutions per site (23). Reference sequences used in the analysis: FCV (NC_001481.2); MCV, mink calicivirus (NC_019712.1); RaVV, rabbit vesivirus (NC_008580.1); SMSV, San Miguel sea lion virus 8 (NC_025676.1), San Miguel sea lion virus 1 (U15301.2); ferret-badger vesivirus (NC_027122.1); SSVV, Steller sea lion vesivirus (NC_011050.1); VESV, vesicular exanthema of swine virus (NC_002551.1); and WCV, walrus calicivirus (NC_004541.1).
TABLE 3.
Percent amino acid identity of CU-296 ORFs to known vesiviruses
| ORF | % amino acid identitya |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FCV CFI/68b | Strain 48 | Strain 2117 | Allston/08 | Allston/09 | Bari/212/07 | Geel/08 | Strain 143 | CU-47 | CU-103 | LSU/96-6931 | |
| ORF1 | 50.0 | 82.6 | 98.9 | 98.8 | 99.0 | 99.3 | 99.4 | ND | ND | ND | ND |
| ORF2 | 41.5 | 71.0 | 93.5 | 87.1 | 86.7 | 87.0 | 88.6 | 87.7 | 89.0 | 84.2 | 94.9 |
| ORF3 | 24.5 | 80.5 | 97.8 | 99.3 | 98.5 | 99.3 | 99.3 | ND | ND | ND | ND |
The highest identities to CU-296 are indicated in boldface. ND, not determined.
FIG 4.
Capsid protein amino acid pairwise distance scan (24) based on the translated nucleotide alignment using a window of 100 nucleotide fragments in 10-nucleotide increments.
TABLE 4.
Percent amino acid identity of the CU-296 capsid to FCV, strain 48, and 2117-like vesivirusesa
| Region | % amino acid identitya |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FCV CFI/68 | Strain 48 | Strain 2117 | Allston/09 | Bari/212/07 | Geel/08 | CU-47 | CU-103 | Strain 143 | LSU/96-6931 | |
| VP1 (full) | 41.5 | 71.0 | 93.5 | 86.7 | 87.0 | 88.6 | 89.0 | 84.2 | 87.7 | 94.9 |
| A, 1–119b | 22.3 | 77.7 | 96.6 | 97.3 | 95.9 | 98.6 | 98.0 | 85.8 | 97.3 | 98.0 |
| B, 120–396 | 53.8 | 80.3 | 96.6 | 90.7 | 91.4 | 91.7 | 92.8 | 92.4 | 91.0 | 97.9 |
| C, 397–411 | 19.0 | 41.7 | 75.0 | 37.5 | 37.5 | 41.7 | 50.0 | 37.5 | 45.8 | 62.5 |
| D, 412–426 | 46.7 | 73.3 | 100 | 73.3 | 73.3 | 80.0 | 86.7 | 73.3 | 86.7 | 100 |
| E, 427–521 | 8.4 | 47.4 | 85.3 | 72.6 | 76.8 | 80.0 | 76.8 | 71.6 | 76.8 | 89.5 |
| F, 522–668 | 39.6 | 66.4 | 93.3 | 88.6 | 86.6 | 87.2 | 88.6 | 85.2 | 86.6 | 95.3 |
The highest identities to CU-296 are indicated in boldface.
Vesivirus strains were aligned to FCV CFI/68 (AAC13993.1) regions with amino acid boundaries as defined by Neill (29).
FIG 5.
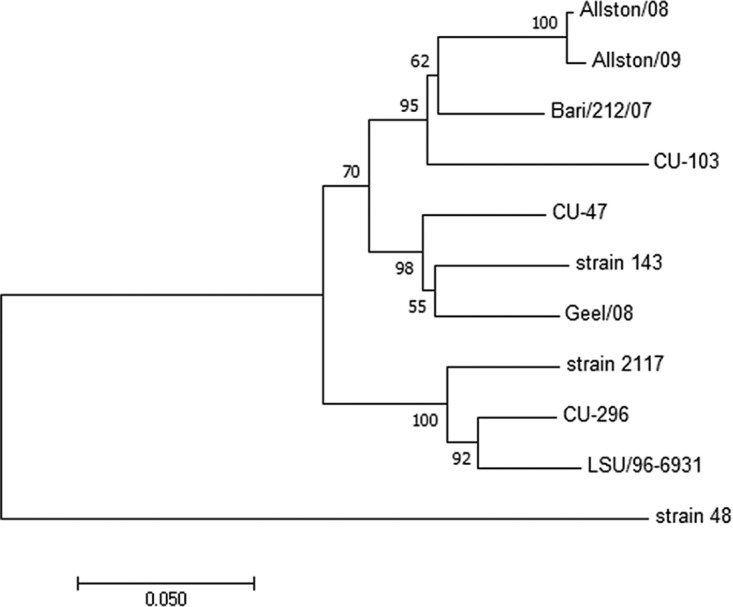
Phylogenetic tree based on the capsid proteins of CaVV and 2117 strains determined using the neighbor-joining method (21). The percentages of 500 replicate trees in which the sequences clustered together are indicated at the nodes (22). Horizontal branch lengths are drawn to the scale of the number of amino acid substitutions per site (23).
RT-PCR assays.
To establish the presence of CaVV in affected dogs, a subset of tissues were tested with three RT-PCR assays specific for CU-296, including FFPE from dogs 1, 2, and 3, and fresh tissues from dog 4 (Table 5). Most FFPE tissues that were tested from dogs 1, 2, and 3 were positive with all three assays. However, kidney tissues from dog 1 and pancreas tissues from dog 2 were negative, and lung tissues from dog 2 and kidney tissues from dog 3 were positive with only two of three and one of three assays, respectively. This may indicate less extensive infection in dogs 1, 2, and 3, but another plausible explanation is a reduced sensitivity of PCR detection with FFPE specimens.
TABLE 5.
Results of RT-PCR screening of FFPE and unfixed tissues
| Dog | Tissue | Assay resulta |
||
|---|---|---|---|---|
| A | B | C | ||
| 1 | Colon | + | + | + |
| Duodenum | + | + | + | |
| Ileum | + | + | + | |
| Jejunum | + | + | + | |
| Kidney | – | – | – | |
| 2 | Lymph node | + | + | + |
| Intestine | + | + | + | |
| Pancreas | – | – | – | |
| Lung | + | – | + | |
| 3 | Abdominal fluid (unfixed) | + | + | + |
| Lung | + | + | + | |
| Duodenum | + | + | + | |
| Ileum | + | + | + | |
| Jejunum | + | + | + | |
| Cerebrospinal fluid (unfixed) | + | + | + | |
| Kidney | – | – | + | |
| 4 | Brain (unfixed) | + | + | + |
| Lung (unfixed) | + | + | + | |
| Colon, small intestine pool (unfixed) | + | + | + | |
The assays are listed in Table S1 in the supplemental material.
Tissue distribution of CaVV nucleic acids.
To further establish the systemic nature of the infection, histological assessment of selected FFPE tissues by ISH with a CaVV-specific probe revealed strong signal localized to the nuclei and cytoplasm of endothelial cells of arterial and capillary blood vessels of nearly every tissue from dogs 1 to 4 (Fig. 1D and Fig. 2A to D and F). Occasional individual leukocytes in the lumen of blood vessels also showed CaVV nucleic acids (Fig. 2F). Consistent with the most severe histological lesions on H&E-stained tissues, all sections of the small intestines from dogs 1, 2, and 4 and of the brain from dog 4 showed prominent CaVV nucleic acids in the endothelial cells of arterioles, small and large arteries, and capillary vessels (Fig. 1D and Fig. 2A to D and F). In addition to endothelial cells, scattered mononuclear cells within the subcapsular sinuses and the cortex of lymph nodes taken from dogs 2 and 4 showed CaVV nucleic acids. Sections of lungs from dogs 3 and 4 had the least amount of CaVV nucleic acids, mostly within scattered endothelial cells of small and large pulmonary arteries and a few circulating leukocytes within the alveolar septa. In the sections of small intestine, CaVV nucleic acids were also found in rare mononuclear cells within the lamina propria at the bases of intestinal crypts. ISH of tissues from dog 4 further extended and confirmed the results seen in dogs 1 to 3. CaVV nucleic acid was widespread within endothelial cells of nearly every arterial and capillary blood vessel of the brain (Fig. 2D), heart, lung, liver (Fig. 2B), spleen (Fig. 2C), kidney, urinary bladder, tongue, stomach, small and large intestine, pancreas, lymph node, dermis, thyroid gland, and cortex of the adrenal gland of dog 4. In the brain of dog 4, CaVV nucleic acids were found in nearly all the endothelial cells of arterial blood vessels and capillaries within the gray and white matter, meninges, and choroid plexus (Fig. 2D and F) but also in scattered glial cells and neurons in the gray matter of the cerebrum and granular cell layer of the cerebellum (Fig. 2D). Besides endothelial cells of arterial blood vessels in the liver of dog 4, Kupffer cells along sinusoids of hepatic lobules showed strong CaVV hybridization (Fig. 2B). Similarly, in the spleen, individual macrophages throughout the red pulp showed strong CaVV signal (Fig. 2C). In the kidney, large arteries, and interstitial capillaries, primarily at the junction between the cortex and medulla, showed a strong hybridization signal. In the bone marrow, rare individual mononuclear cells showed CaVV signal. ISH of a small intestinal section from dog 1 with control probes Dap-B and CI-UBC showed negative and positive reactivity, respectively. As expected, staining of control uninfected and CaVV-infected CK-DE2 cells confirmed the specificity of the CaVV probe, with 90% of infected cells showing strong cytoplasmic signal, while uninfected cells were negative.
DISCUSSION
A focal outbreak of hemorrhagic gastroenteritis occurred over a period of several weeks in a well-managed, high-volume, pet boarding facility in a major metropolitan area. Enhanced biosecurity measures effectively controlled the outbreak, and spread of the illness to other boarding facilities or domestic settings was not reported. The suspicion of a viral cause and no known etiology that could result in mortality within 24 h of onset of clinical signs led to an expanded search for an unknown pathogen. We propagated a 2117-like vesivirus from fresh tissue obtained from dog 4 and confirmed the virus by RT-PCR in the FFPE tissues from the other three dogs. ISH was used to study the extent of infection and tissue tropism. On the basis of ISH with a probe specific for CaVV, deceased dogs 1 to 4 showed systemic spread primarily within endothelial cells of arterial and capillary blood vessels that likely accounts for the rapid clinical progression of the disease and the demise of these dogs. Consistent with the distribution of CaVV nucleic acids by ISH, fibrinoid necrosis of arterial walls and thrombosis of capillary blood vessels suggested ischemia secondary to vascular injury as the underlying mechanism for the extensive intestinal mucosal necrosis and hemorrhage seen in dogs 1, 2, and 4. Interestingly, the distribution of CaVV nucleic acids targeting endothelial cells mirrors Shiga toxin-mediated vascular damage resulting in hemorrhagic colitis and hemolytic-uremic syndrome in humans infected with enteropathogenic Escherichia coli strains (31). In addition to vascular endothelium, CaVV nucleic acids were found in scattered circulating leukocytes in the lumina of blood vessels, scattered microglial cells, and rare neurons in the cerebrum and cerebellum, occasional mononuclear cells in the lamina propria of the intestine, red pulp of the spleen, sinus and cortex of the lymph node, and bone marrow. The significance of these findings is uncertain but suggests that cells other than endothelial cells are infected with CaVV. The tropism of CaVV for endothelial cells and the pattern of disease characterized by systemic infection are analogous to rare vsFCV epizootics and support the suggestion of a hypervirulent variant CaVV as the cause of this disease outbreak. The only significant host inflammatory and immune response to CaVV was seen in the brain of dog 4 and was characterized by a mild lymphohistiocytic perivascular infiltrate consistent with a nonsuppurative meningoencephalitis and choroid plexitis with multifocal gliosis. Given that the majority of nonsuppurative meningoencephalitides of dogs have an unknown etiology, CaVV might account for a subset of these previously undiagnosed cases (32).
Feline caliciviruses are typically associated with respiratory disease or oral lesions and are not thought of as a cause of enteric signs. Clinical signs with field strains of FCV can range broadly in severity and include vesicular lesions on the tongue or palate with mildly virulent strains, progressing to fever, pneumonia, and depression with highly virulent strains; however, mortality is rare (33). Clinical findings in cats infected with vsFCV are consistent with those of low-virulence strains, but they are more severe and may most notably result in up to 50% mortality (3). The hemorrhagic-like fever described in the first outbreak (34) has not been observed in subsequent vsFCV outbreaks quite possibly indicating a range of virulence among vsFCV strains. The mechanism of disease and mortality remains undetermined but an immune-mediated cytokine response has been suggested (35). Systemic infection is common to both vsFCV and CU-296, but there are clear distinctions in the clinical profiles. Respiratory disease was not a presenting concern in any of the 11 dogs in our study, while hemorrhagic gastroenteritis and acute vomiting were common to all dogs. In experimental infections of vsFCV, antigen is associated with epithelial cell damage in the respiratory tract, skin, and mucosa and in vascular endothelial cells of the dermis (36). Both vsFCV and CU-296 target endothelial cells, but in our study CaVV preferentially targets endothelial cells of arterial and capillary blood vessels.
Amino acid differences or other features unique to CU-296 that could lead to a hypervirulent phenotype reminiscent of vsFCV were not identified by comparative analysis. Previous efforts to identify unique determinants in vsFCV strains focused on hypervariable regions of the capsid protein, but researchers were not able to point to specific genetic differences that were not also present in some common and presumably lower-virulence strains (35, 37, 38). The significant variation observed in CaVV VP1 sequences may mean that elucidating the changes necessary to go from low to high virulence may be equally challenging. Cryo-EM structures of 2117 virus-like particles have revealed that at least structurally the 2117 strains more closely resemble sapoviruses and rabbit hemorrhagic disease virus (Lagovirus) than other members of the vesivirus genus (39). Comparison to the structure of FCV, which uses junctional adhesion molecule-A (40) or α-2,6 sialic acid (41), reveals significant differences in the P2 binding domains, leading to speculation that 2117 strains may not use either of these receptors. It is also possible that the capsid sequence is not the most important genetic determinant of the hypervirulent systemic phenotype.
Serological surveys provide convincing evidence of endemic infections of dogs with CaVV. The earliest work showed that 76% of the sera (n = 125) from Tennessee were positive by IFA (10) and that 57% were positive (n = 244) to strain 48 by neutralization in Japan (42) and 36.5% (n = 319) in South Korea (43). Most recently, antibodies to 2117-like strains have been confirmed in 21.5% of domestic dogs by enzyme-linked immunosorbent assay (44). Our preliminary efforts to identify antibodies to CU-296 in dogs based on serum neutralization (SN) assays have been inconsistent with previous findings (titers of 1:4 to 1:24 in 8/178 dogs). Marine vesivirus and FCV strains commonly evade cross-neutralization by nonhomologous serum (45, 46) due to variation mapped to the E hypervariable region in FCV VP1 (47, 48). The consistent high variability in the region corresponding to FCV E indicates that SN assays might not provide a robust assessment of CaVV prevalence. The finding of two additional CaVV strains in dogs of undetermined health status supports the argument for endemic infection but does not provide conclusions about an etiological association with gastrointestinal disease. The observed prevalence of 1.4% is consistent with the 1.1 and 3.5% reported from Italy in both gastrointestinal disease and healthy dogs, respectively (17). Strain 143 was obtained from archived stocks, and information was limited to the vial label identifying it as “canine calicivirus 1997.” The research pertaining to CaVV isolated from genital lesions dates from a decade earlier, so it cannot be assumed that strain 143 is one of the isolates originally described (11). LSU/96-6931 was isolated from the brain tissue of a dog with meningoencephalitis in 1996 and identified as a calicivirus-like virus by TEM at the time. Archival tissues were not available to confirm the systemic nature of this infection; however, neurological signs suggest a disease pattern similar to that seen in dog 4 of the present study.
Indeterminate clinical findings in the dogs in the outbreak and the identification of an uncommon agent provides a plausible argument for CU-296 as the cause of the hemorrhagic gastroenteritis and systemic spread that proved fatal. A conclusive determination will require reproduction of the disease syndrome in pathogen-free dogs. It is also important to determine the origin of 2117 strains in biopharmaceutical and laboratory cultures. Initially, 2117 group strains were known to replicate only in CHO cells. The initial 2117 isolate did not replicate in Vero, BHK-21, MDCK, Madin-Darby bovine kidney, Crandell feline kidney, porcine kidney 15, HeLa, or CaCo2 cells (14). However, in addition to the culture of CU-296 in two canine cell lines in our study, Allston/2008 has since been cultured in BHK-21 and MDCK cells (49), and strain 143 was originally cultured in MDCK and dolphin kidney cells (11). The ability of these viruses to replicate in cell lines from diverse host species suggests additional potential reservoirs and transfer to susceptible dog populations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nancy Zylich and Erica Butler for technical assistance.
This study was supported by development funds from the Animal Health Diagnostic Center, College of Veterinary Medicine, Cornell University.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01951-17.
REFERENCES
- 1.Smith AW, Boyt PM. 1990. Caliciviruses of ocean origin: a review. J Zoo Wildl Med 21:3–23. [Google Scholar]
- 2.Smith AW, Skilling DE, Cherry N, Mead JH, Matson DO. 1998. Calicivirus emergence from ocean reservoirs: zoonotic and interspecies movements. Emerg Infect Dis 4:13–20. doi: 10.3201/eid0401.980103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pesavento PA, Chang KO, Parker JS. 2008. Molecular virology of feline calicivirus. Vet Clin North Am Small Anim Pract 38:775–786. doi: 10.1016/j.cvsm.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Evermann JF, Bryan GM, Mckeirnan A. 1981. Isolation of a calicivirus from a case of canine glossitis. Canine Pract 8:36–39. [Google Scholar]
- 5.Evermann JF, Mckeirnan AJ, Smith AW, Skilling DE, Ott RL. 1985. Isolation and identification of caliciviruses from dogs with enteric infections. Am J Vet Res 46:218–220. [PubMed] [Google Scholar]
- 6.Gabriel S, Tohya Y, Mochizuki M. 1996. Isolation of a calicivirus antigenically related to feline caliciviruses from feces of a dog with diarrhoea. J Vet Med Sci 58:1041–1043. doi: 10.1292/jvms.58.10_1041. [DOI] [PubMed] [Google Scholar]
- 7.Pratelli A, Greco G, Camero M, Normanno G, Buonavoglia C. 2000. Isolation and identification of a calicivirus from a dog with diarrhoea. New Microbiol 23:257–260. [PubMed] [Google Scholar]
- 8.Martella V, Pratelli A, Gentile M, Buonavoglia D, Decaro N, Fiorante P, Buonavoglia C. 2002. Analysis of the capsid protein gene of a feline-like calicivirus isolated from a dog. Vet Microbiol 85:315–322. doi: 10.1016/S0378-1135(01)00521-1. [DOI] [PubMed] [Google Scholar]
- 9.Di Martino B, Di Rocco C, Ceci C, Marsilio F. 2009. Characterization of a strain of feline calicivirus isolated from a dog faecal sample. Vet Microbiol 139: 52–57. doi: 10.1016/j.vetmic.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaffer FL, Soergel ME, Black JW, Skilling DE, Smith AW, Cubitt WD. 1985. Characterization of a new calicivirus isolated from feces of a dog. Arch Virol 84:181–195. doi: 10.1007/BF01378971. [DOI] [PubMed] [Google Scholar]
- 11.Crandell RA. 1988. Isolation and characterization of caliciviruses from dogs with vesicular genital disease. Arch Virol 98:65–71. doi: 10.1007/BF01321006. [DOI] [PubMed] [Google Scholar]
- 12.Mochizuki M, Kawanishi A, Sakamoto H, Tashiro S, Fujimoto R, Ohwaki MA. 1993. Calicivirus isolated from a dog with fatal diarrhoea. Vet Rec 132:221–221. doi: 10.1136/vr.132.9.221. [DOI] [PubMed] [Google Scholar]
- 13.Roerink F, Hashimoto M, Tohya Y, Mockizuki M. 1999. Genetic analysis of canine calicivirus: evidence for a new clade of animal caliciviruses. Vet Microbiol 69:69–72. doi: 10.1016/S0378-1135(99)00091-7. [DOI] [PubMed] [Google Scholar]
- 14.Oehmig A, Buttner M, Weiland F, Werz W, Bergemann-Pfaff KE. 2003. Identification of a calicivirus isolate of unknown origin. J Gen Virol 84:2837–2845. doi: 10.1099/vir.0.19042-0. [DOI] [PubMed] [Google Scholar]
- 15.Allison M. 2010. As Genzyme flounders, competitors and activist investors swoop. Nat Biotechnol 28:3–4. doi: 10.1038/nbt0110-3. [DOI] [PubMed] [Google Scholar]
- 16.Castro TX, Cubel-Garcia RCN, Costa EM, Leal RM, da Xavier PTM, Leite JPG. 2013. Molecular characterization of calicivirus and astrovirus in puppies with enteritis. Vet Rec 172:557–559. doi: 10.1136/vr.101566. [DOI] [PubMed] [Google Scholar]
- 17.Martella V, Pinto P, Lorusso E, Di Martino B, Wang Q, Larocca V, Cavalli A, Comero M, Decaro N, Banyai K, Saif L, Buonavoglia C. 2015. Detection and full-length genome characterization of novel canine vesiviruses. Emerg Infect Dis 21:1433–1436. doi: 10.3201/eid2108.140900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreier J, Störmer M, Kleesiek K. 2005. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J Clin Microbiol 43:4551–4557. doi: 10.1128/JCM.43.9.4551-4557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmonds P. 2012. SSE: a nucleotide and amino acid sequence analysis platform. BMC Res Notes 5:50. doi: 10.1186/1756-0500-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed Z, Shaw G, Sharma VP, Yang C, McGowan E, Dickson DW. 2007. Actin-binding proteins Coronin-1a and IBA-1 are effective microglial markers for immunohistochemistry. J Histochem Cytochem 55:687–700. doi: 10.1369/jhc.6A7156.2007. [DOI] [PubMed] [Google Scholar]
- 26.Jager MC, Sloma EA, Shelton M, Miller AD. 2017. Naturally acquired canine herpesvirus-associated meningoencephalitis. Vet Pathol 54:820–827. doi: 10.1177/0300985817716263. [DOI] [PubMed] [Google Scholar]
- 27.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. 2012. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO. 1999. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J Virol Methods 83:145–154. doi: 10.1016/S0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- 29.Neill JD. 1992. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among calicivirus capsid proteins. Virus Res 24:211–222. doi: 10.1016/0168-1702(92)90008-W. [DOI] [PubMed] [Google Scholar]
- 30.Neill JD, Meyer RF, Seal BS. 1998. The capsid protein of vesicular exanthema of swine virus serotype A48: relationship to the capsid protein of other animal caliciviruses. Virus Res 54:39–50. doi: 10.1016/S0168-1702(98)00013-6. [DOI] [PubMed] [Google Scholar]
- 31.Pruimboom-Brees IM, Morgan TW, Ackermann MR, Nystrom ED, Samuel JE, Cornick NA, Moon HW. 2000. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc Natl Acad Sci U S A 97:10325–10329. doi: 10.1073/pnas.190329997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwab S, Herden C, Seeliger F, Papaioannou N, Psalla D, Polizopulou Z, Baumgärtner W. 2007. Non-suppurative meningoencephalitis of unknown origin in cats and dogs: an immunohistochemical study. J Comp Pathol 136:96–110. doi: 10.1016/j.jcpa.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoover EA, Kahn DE. 1975. Experimentally induced feline calicivirus infection: clinical signs and lesions. J Am Vet Med Assoc 166:463–468. [PubMed] [Google Scholar]
- 34.Pedersen NC, Elliott JB, Glasgow A, Poland A, Keel K. 2000. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet Microbiol 73:281–300. doi: 10.1016/S0378-1135(00)00183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foley J, Hurley K, Pesavento PA, Poland A, Pedersen NC. 2006. Virulent systemic feline calicivirus infection: local cytokine modulation and contribution of viral mutants. J Feline Med Surg 8:55–61. doi: 10.1016/j.jfms.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesavento PA, MacLachlan NJ, Dillard-Telm L, Grant CK, Hurley KF. 2004. Pathologic, immunohistochemical and electron microscopic finds in naturally occurring virulent systemic feline calicivirus infection in cats. Vet Pathol 41:257–263. doi: 10.1354/vp.41-3-257. [DOI] [PubMed] [Google Scholar]
- 37.Rong S, Slade D, Floyd-Hawkins K, Wheeler D. 2006. Characterization of a highly virulent feline calicivirus and attenuation of this virus. Virus Res 122:95–108. doi: 10.1016/j.virusres.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Ossiboff RJ, Sheh A, Shotton J, Pesavento PA, Parker JS. 2007. Feline caliciviruses (FCVs) isolated from cats with virulent systemic disease possess in vitro phenotypes distinct from those of other FCV isolates. J Gen Virol 88:506–517. doi: 10.1099/vir.0.82488-0. [DOI] [PubMed] [Google Scholar]
- 39.Conley M, Emmott E, Orton R, Taylor D, Carneiro DG, Murata K, Goodfellow IG, Hansman GS, Bhella D. 2017. Vesivirus 2117 capsids more closely resemble sapovirus and lagovirus particles than other known vesivirus structures. J Gen Virol 98:68–76. doi: 10.1099/jgv.0.000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makino A, Shimojima M, Miyazawa T, Kato K, Tohya Y, Akashi H. 2006. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J Virol 80:4482–4490. doi: 10.1128/JVI.80.9.4482-4490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stuart AD, Brown TD. 2007. Alpha2,6-linked sialic acid acts as a receptor for Feline Calicivirus. J Gen Virol 88:177–186. doi: 10.1099/vir.0.82158-0. [DOI] [PubMed] [Google Scholar]
- 42.Mochizuki M, Hashimoto M, Roerink F, Tohya Y, Matsuura Y, Sasaki N. 2002. Molecular and seroepidemiological evidence of canine calicivirus infections in Japan. J Clin Microbiol 40:269–2631. doi: 10.1128/JCM.40.7.2629-2631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang HK, Tohya Y, Han KY, Kim TJ, Song CS, Mochizuki M. 2003. Seroprevalence of canine calicivirus and canine minute virus in the Republic of Korea. Vet Rec 153:150–152. doi: 10.1136/vr.153.5.150. [DOI] [PubMed] [Google Scholar]
- 44.Di Martino B, Di Profio F, Bodnar L, Melegari I, Sarchese V, Massirio I, Dowgier G, Lanave G, Marsilio F, Bányai K, Buonavoglia C, Martella V. 2017. Seroprevalence for 2117-like vesiviruses in Italian household dogs. Vet Microbiol 201:14–17. doi: 10.1016/j.vetmic.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Smith AW, Latham AB. 1978. Prevalence of vesicular exanthema of swine antibodies among feral mammals associated with the southern California coastal zones. Am J Vet Res 39:291–296. [PubMed] [Google Scholar]
- 46.Radford AD, Bennett M, McArdle F, Dawson S, Turner PC, Glenn M, Gaskell RM. 1997. The use of sequence analysis of a feline calicivirus (FCV) hypervariable region in the epidemiological investigation of FCV related disease and vaccine failures. Vaccine 15:1451–1458. doi: 10.1016/S0264-410X(97)00059-5. [DOI] [PubMed] [Google Scholar]
- 47.Neill JD, Sosnovtsev SV, Green KY. 2000. Recovery and altered neutralization specificities of chimeric viruses containing capsid protein domain exchanges from antigenically distinct strains of feline calicivirus. J Virol 74:1079–1084. doi: 10.1128/JVI.74.3.1079-1084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geissler K, Schneider K, Truyen U. 2002. Mapping neutralizing and non-neutralizing epitopes on the capsid protein of feline calicivirus. J Vet Med B Infect Dis Vet Public Health 49:55–60. doi: 10.1046/j.1439-0450.2002.00529.x. [DOI] [PubMed] [Google Scholar]
- 49.Plavsic M, Shick K, Bergmann F, Mallet L. 2016. Vesivirus 2117: cell line infectivity range and effectiveness of amplification of a potential adventitious agent in cell culture used for biological production. Biologicals 44:540–545. doi: 10.1016/j.biologicals.2016.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.