Fig. 2.
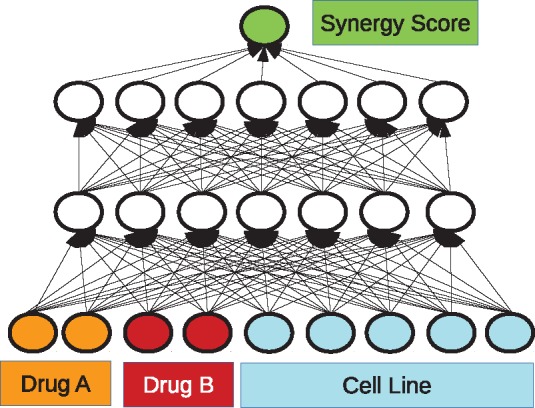
Schematic illustration of our Deep Learning approach. The input consists of three parts: the chemical descriptors for drug A and drug B, and the genomic information of the cell line. The inputs are propagated through the network to the linear output unit. The thereby obtained result is the predicted synergy value. The best performing architecture was determined via exploration of different hyperparameters which are listed in Table 1
