Fig. 3.
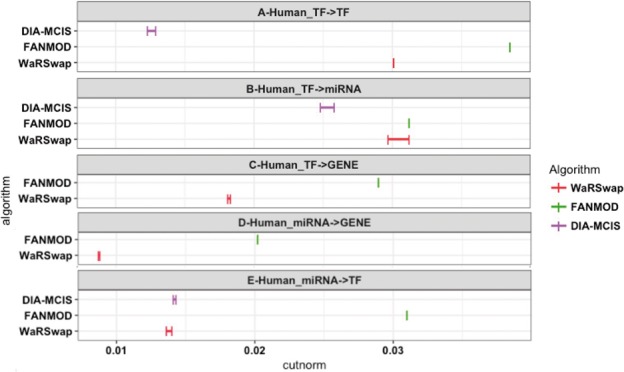
Human TF-miRNA-Gene network sampling performance. A total of 5000 graphs were generated by each algorithm and the cut norm estimates were computed using IndeCut. The vertical lines represent lower and upper bounds returned by the cut norm estimation with the true (NP-hard) value lying in this interval. A cut norm interval that is far from zero represents less uniform and independent sampling. The cut norm estimates for CoMoFinder were much larger than 0.04, and hence were removed for ease of comparison. In panels C and D, results for DIA-MCIS are absent since this algorithm does not operate on graphs with more than 2035 nodes (see Table S1 and Supplementary Fig. S10 for detailed results)
