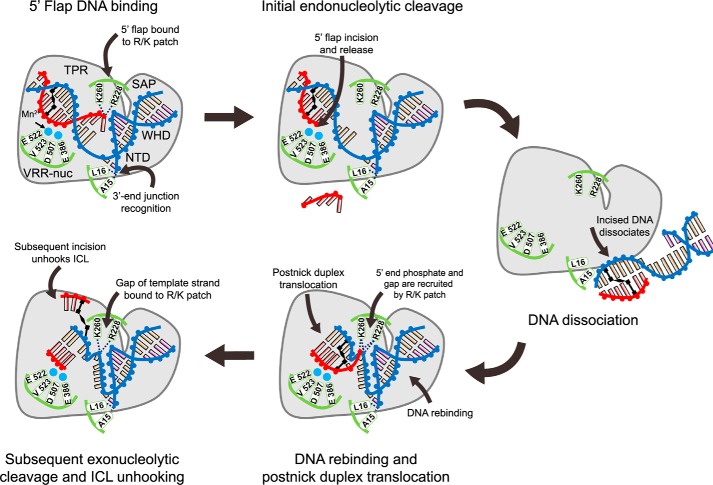
Figure 7.
Model for the ICL unhooking by PaFAN1. DNA strands are labeled as blue (template strand and pre-nick strand), red (5′ flap strand), and black for ICL-coupled bases. Mn2+ ions are shown in cyan spheres, and key residues for DNA recognition and cleavage are in green. (i) Recognition of a DNA substrate with a 5′ flap and an ICL by PaFAN1: pre-nick duplex by NTD, post-nick duplex by CTD, 3′-end junction by N-terminal loop, and 5′ flap by the Arg/Lys patch; (ii) 5′ flap is removed by initial endonucleolytic cleavage; (iii) the product is released from PaFAN1; (iv) PaFAN1 rebounds to the cleaved fragment and locates the next scissile phosphate group at the active site through the recognition of the phosphate of a gap by a Arg/Lys patch; (v) Subsequent exonucleolytic cleavages eventually unhook an ICL. The key features of PaFAN1 for ICL unhooking are marked: (a) 5′-terminal phosphates by a Arg/Lys patch (Arg-228 and Lys-260), (b) the pre-nick region by the N-terminal loop, and (c) a phosphate group at the gap by a Arg/Lys patch.