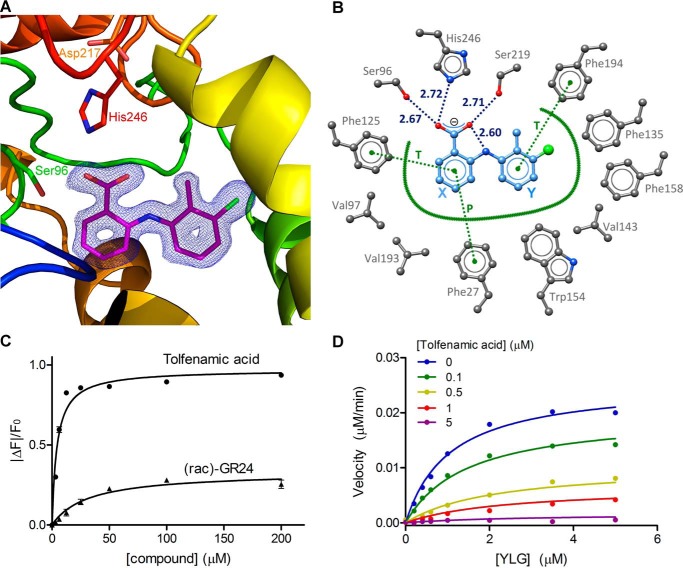
Figure 4.
DAD2 inhibition by tolfenamic acid. A, structure of DAD2 bound to tolfenamic acid. DAD2 is drawn in ribbon mode and rainbow-colored from blue (N terminus) to red (C terminus). The catalytic triad residues and tolfenamic acid (pink) are shown in stick mode. The final σA-weighted map contoured at 1.3σ around tolfenamic acid is shown in dark blue (the corresponding omit map is shown in Fig. S2). B, tolfenamic acid binding mode within DAD2's internal cavity. Oxygen, nitrogen, and chlorine atoms are represented in red, blue, and green spheres, respectively. Carbon atoms are shown as gray spheres for protein atoms, and light blue spheres for tolfenamic acid. Hydrogen bonds are shown as dotted blue lines, with distances (in Å) between polar atoms indicated. Hydrophobic interactions are represented by a thick green line. Specific π–π stacking interactions are shown as dotted green lines and labeled T and P for perpendicular T-stack and parallel stack, respectively. Residue numbers are indicated in gray. Ring labels (X and Y) used throughout are indicated in blue. C, intrinsic fluorescence of DAD2 in the presence of tolfenamic acid and rac-GR24. Each data point is the mean ± S.E. (error bars) of three technical replicates. D, competition assay of YLG hydrolysis by DAD2 using tolfenamic acid. Each data point is the average of three technical replicates. All of the individual replicates for each compound concentration were included during the nonlinear global fit analysis using a mixed-inhibition model. See also Table S2.