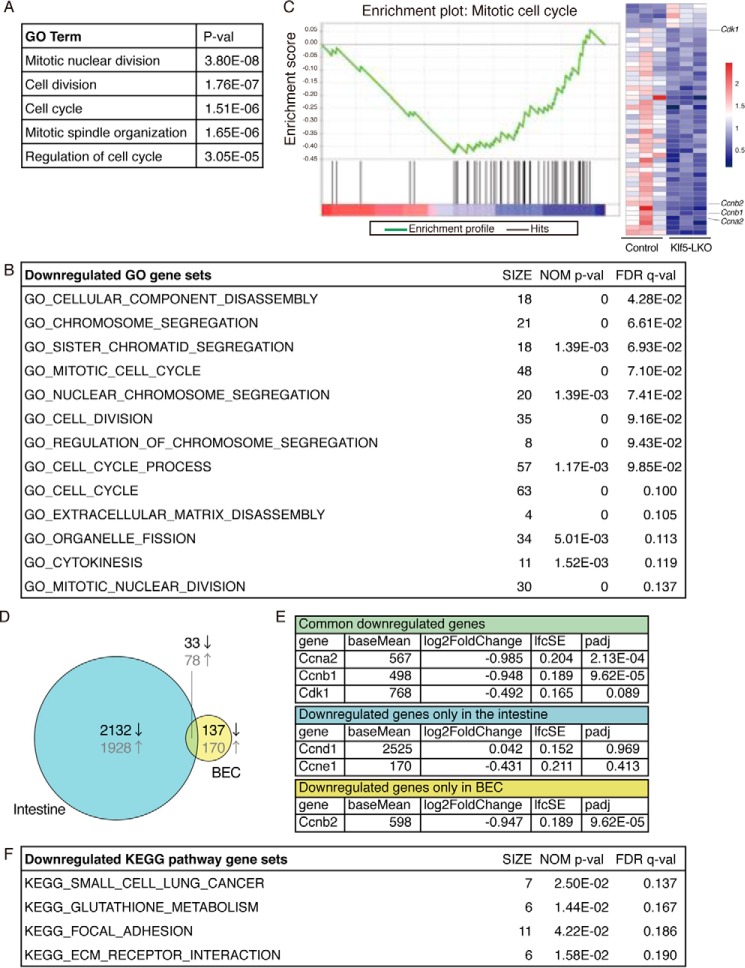
Figure 5.
Transcriptome analysis by RNA-seq reveals that Klf5 regulates cell proliferation in BECs. A, DEG were categorized in biological GO terms using DAVID. B and C, GSEA using DEG to identify enriched biological GO terms. B, entire list of enriched gene sets categorized in GO biological process terms that were down-regulated by Klf5 deletion in BECs. The SIZE column indicates the number of genes hit in each gene set. Gene sets that meet the criteria for both NOM p value of <0.05 and FDR q value of <0.25 are considered to be significantly enriched and are listed. C, representative enrichment plot, corresponding to the “GO_MITOTIC_CELL_CYCLE” set in B. The heat map shows expression levels of genes included in the gene set. The left three and right three columns correspond to BEC samples from the control and Klf5-LKO livers, respectively. The expression levels are indicated according to the scale bar at right. D, Venn diagram showing DEG identified in the intestine (left circle) and BECs (right circle) upon Klf5 deletion. Numbers shown in black and gray characters in the diagram indicate the counts of down-regulated and up-regulated genes, respectively. E, RNA-seq data for expression levels for cell cycle-related genes. Three categories correspond to those in the Venn diagram shown in D. F, entire list of enriched KEGG pathway gene sets. The SIZE column indicates the number of genes hit in each gene set. Gene sets that meet the criteria for both NOM p value <0.05 and FDR q value <0.25 are considered to be significantly enriched and are listed.