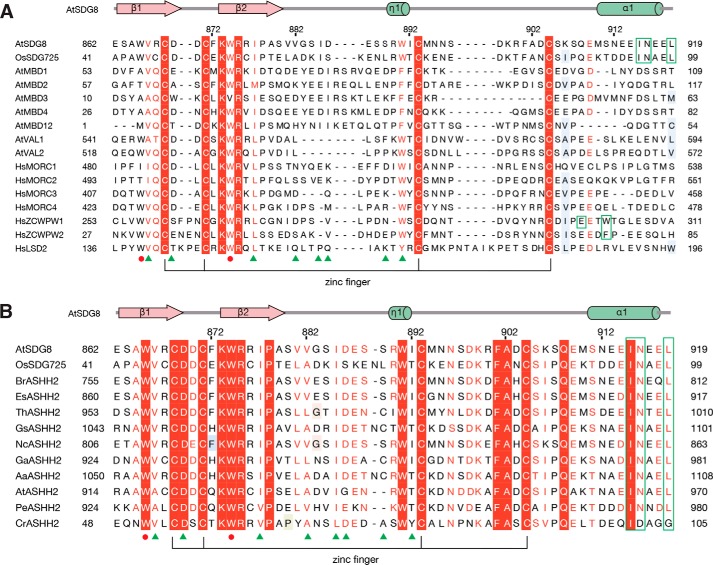
Figure 5.
Sequence alignment of CW proteins in species. Sequence alignment was calculated using Jalview (53), and amino acids were shaded according to the ESPript server (54). Secondary structural elements of SDG8-CW are displayed above the sequence alignment. The conserved zinc-binding mode is shown by lines at the bottom of the alignment. The residues that form the conserved aromatic cage are marked by red circles, and the various residues of the aromatic cage are marked by green frames. The key residues involved in histone tail sequence-specific recognition are marked by green triangles. A, sequence alignment of CW domains in human (Hs) and plant (Os and At). B, sequence alignment of SDG8 CW domain with its homologs from other plants. Br, Brassica rapa, XP_018508679.1; Es, Eutrema salsugineum, XP_006390102.1; Th, Tarenaya hassleriana, XP_010534709.1; Gs, Glycine soja, KHN44299.1; Nc, Noccaea caerulescens, JAU85503.1; Ga, Gossypium arboreum, XP_017641601.1; Aa, Anthurium amnicola, JAT51201.1; At, Aegilops tauschii, EMT20452.1; Pe, Phalaenopsis equestris, XP_020573738.1; Cr, Chlamydomonas reinhardtii, XP_001694743.1.