Fig. 1.
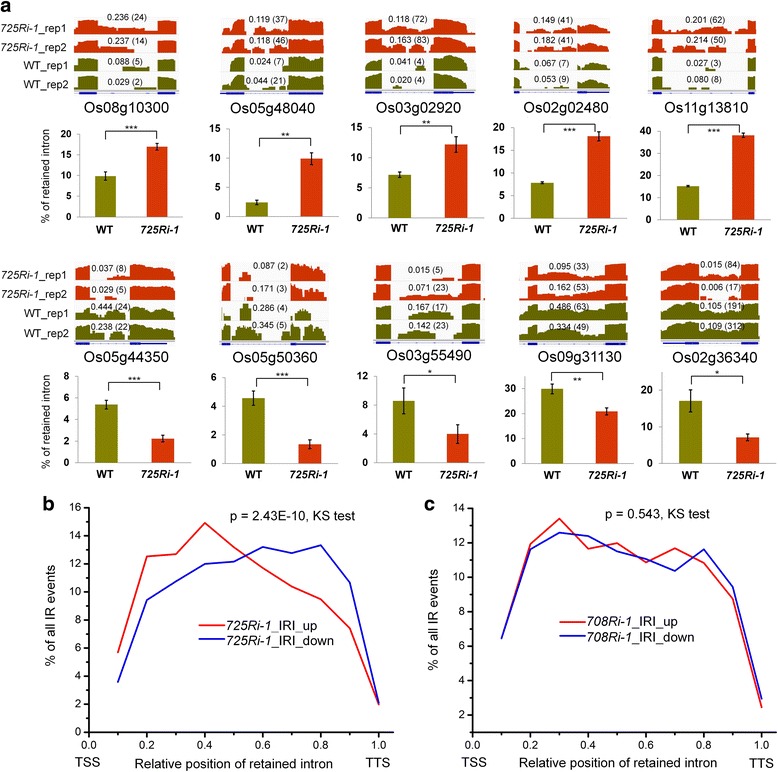
SDG725, but not SGD708, affects global intron retention shift. a RNA-seq tracks (log2 transformed) and qRT-PCR validation for 10 differentially retained introns between 725Ri-1 and WT plants. Two biological replicates (rep1 and rep2) of RNA-seq data of 725Ri-1 and WT are shown. IRI and read numbers (in parentheses) are shown in the corresponding introns. Rice genes are shown below RNA-seq tracks. *, **, and *** indicate p values < 0.05, < 0.01, and < 0.001, respectively (t test). b, c Distribution of up-regulated (red) and down-regulated (blue) intron retention (IR) events between 725Ri-1 and wild-type (WT) rice (b), and between 708Ri-1 and WT (c). A twofold change in IRI was applied as a cutoff to define differential IR events. The retained introns also must be supported by at least 3 reads, length coverage ≥80%, and host gene expression ≥1 FPKM. Average IRI value of two biological replicates of both WT and mutant was used for location distribution Kolmogorov-Smirnov (KS) test. TSS transcription start site, TTS transcription termination site
