Figure 2.
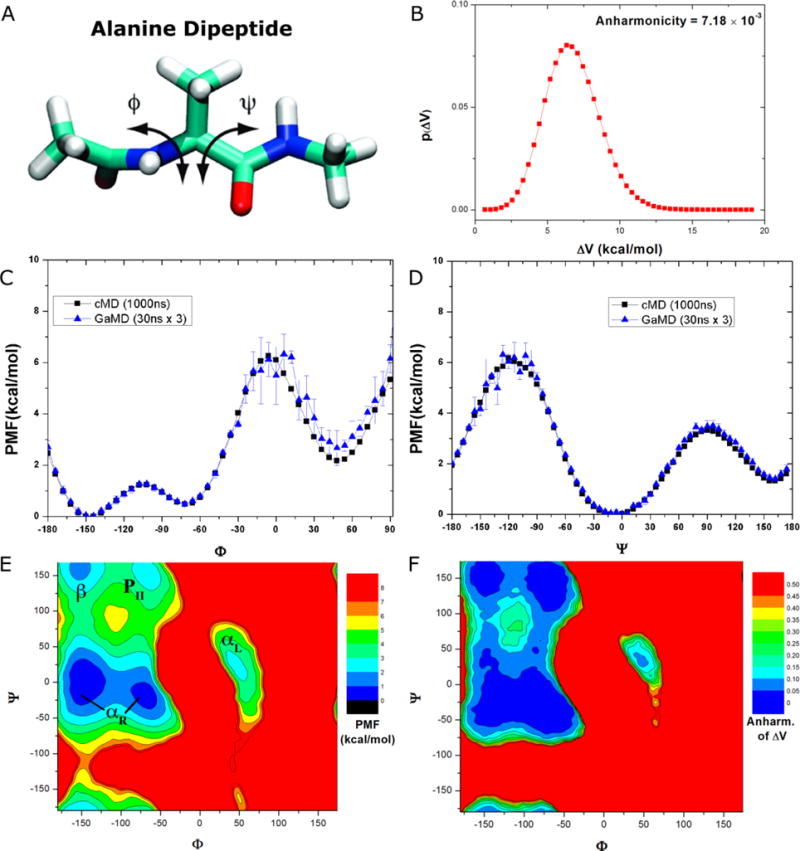
Demonstration of GaMD on the alanine dipeptide: (A) Schematic representation of backbone dihedrals Φ and Ψ in alanine dipeptide. (B) Distribution of the boost potential applied in the GaMD simulations with anharmonicity equal to 7.18×10−3. (C-D) potential of mean force (PMF) profiles of the (C) Φ and (D) Ψ dihedrals calculated from three 30 ns GaMD simulations combined using cumulant expansion to the 2nd order. (E) The 2D PMF profile of backbone dihedrals (Φ, Ψ). The low energy wells are labeled corresponding to the right-handed α helix (αR), left-handed α helix (αL), β-sheet (β) and polyproline II (PII) conformations. (F) The distribution anharmonicity of of frames found in each bin of the PMF profile.
