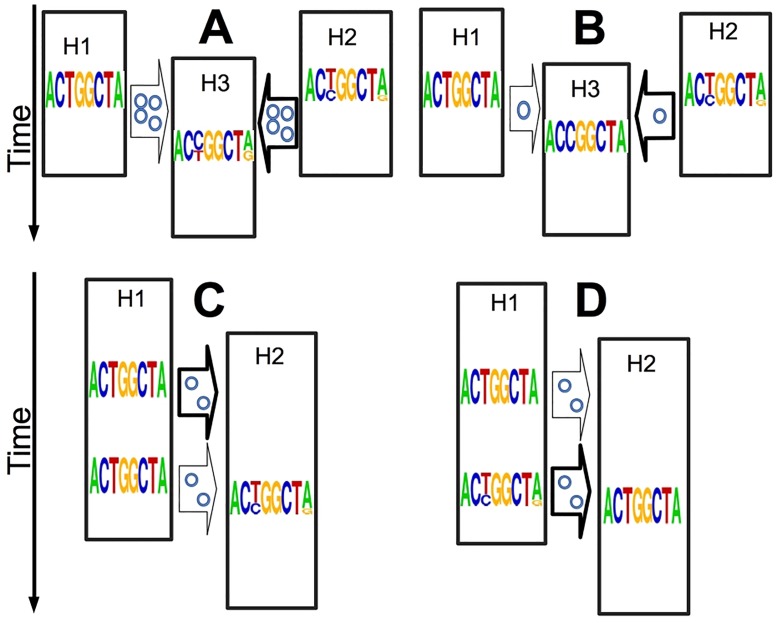
Fig 1. Examples of informativeness of within-host genetic variants.
Here we show how within-host within-sample genetic variants can be useful without requiring pathogen haplotypes. Each string of letters (a frequency sequence logo [34, 35]) represents the collective genome of the pathogen at a certain point in time, as could be observed through deep sequencing. Multiple letters in the same column represent a genetic variant, with letter size representing allelic abundance. Time is on the Y axis, hosts are represented as black rectangles (a host is only active in the outbreak for the portion of vertical axis it occupies), and plausible transmission events as arrows. The posterior probability of different transmission events is represented by the arrow thickness. The number of little circles within arrows represents the inoculum size (transmission bottleneck). A) Shared genetic variants hint to epidemiological relatedness: the two top hosts (H1 and H2) are both possible infectors of the central host (H3), but H2 shares two genetic variants with H3, making it a likely infector of H3. Furthermore, the presence of shared genetic variants suggests a large transmission inoculum (a weak transmission bottleneck). B) A genetic variant of the same type of a substitution can hint to an infector: as before, but now H3 has a substitution (at third genome position, from T to C), which means that its within-host population is non-polymorphic at this position, but with a different nucleotide than the index case. This substitution is between the two nucleotides present at the same position in H2 (where this position is a genetic variant), consistent with H2 being the infector of H3. Also, this time the absence of shared genetic variants is indicative of a small transmission inoculum (a strong transmission bottleneck). C-D) The number of new genetic variants is informative of the age of an infection (but possibly also of the history of the pathogen population size within the host): in C the presence of non-shared variants in H2 suggests that the infection is older, while in D their absence suggests that the infection is younger.