SUMMARY
The aminoacyl-tRNA synthetases and their cognate transfer RNAs translate the universal genetic code. The twenty canonical amino acids are sufficiently diverse to create a selective advantage for dividing amino acid activation between two distinct, apparently unrelated superfamilies of synthetases, Class I amino acids being generally larger and less polar, Class II amino acids smaller and more polar. Biochemical, bioinformatic, and protein engineering experiments support the hypothesis that the two Classes descended from opposite strands of the same ancestral gene. Parallel experimental deconstructions of Class I and II synthetases reveal parallel losses in catalytic proficiency at two novel modular levels—protozymes and Urzymes—associated with the evolution of catalytic activity. Bi-directional coding supports an important unification of the proteome; affords a genetic relatedness metric—middle base-pairing frequencies in sense/antisense alignments—that probes more deeply into the evolutionary history of translation than do single multiple sequence alignments; and has facilitated the analysis of hitherto unknown coding relationships in tRNA sequences. Reconstruction of native synthetases by modular thermodynamic cycles facilitated by domain engineering emphasizes the subtlety associated with achieving high specificity, shedding new light on allosteric relationships in contemporary synthetases. Synthetase Urzyme structural biology suggests that they are catalytically active molten globules, broadening the potential manifold of polypeptide catalysts accessible to primitive genetic coding and motivating revisions of the origins of catalysis. Finally, bi-directional genetic coding of some of the oldest genes in the proteome places major limitations on the likelihood that any RNA World preceded the origins of coded proteins.
Keywords: Synthetase Class division, Bi-directional genetic coding, Urzymes, Protozymes, Modular deconstruction, reflexivity
I. Introduction
“It is unlikely that the aminoacyl-tRNA synthetases played any specific role in the evolution of the genetic code; their evolutions did not shape the codon assignments.” [1]
“A real understanding of the code origin and evolution is likely to be attainable only in conjunction with a credible scenario for the evolution of the coding principle itself and the translation system.” [2]
The first epigram begins the concluding section of an authoritative review of this field published in 2000. The review contains much information and detailed interpretations based on the best available data at that time. Much of the research and theory to emerge since that time, however, has pointed to the opposite conclusion, in keeping with the dialectic component always implicit in scientific research. Consistent with the spirit expressed in the second epigram, unprecedented experimental and bioinformatic studies of the earliest evolution of aminoacyl-tRNA synthetases (aaRS) now make a compelling case for their intimate and probably necessary participation, with tRNA, in the evolution of the universal genetic code and the shaping of codon assignments.
A. The RNA World Hypothesis
The results reviewed herein are especially relevant to the question of whether or not present day biology replaced a prior organization in which information storage and catalysis both were entirely the province of RNA [3,4]. As argued elsewhere in detail [5,6], the actual evidence for such scenarios is remarkably thin. The proposal that aminoacyl-tRNA synthetase enzymes arose as a single pair of ancestors coded bi-directionally on opposite strands of the same RNA gene decisively undermines the heart of the RNA World scenario, by establishing that catalysis of aminoacylation by proteins emerged with scarcely non-random fidelity. Such lack of specificity would have been abolished by purifying selection, had there been any ribozymal system with higher fidelity.
Moreover, aminoacyl-tRNA synthetases, aaRS, represent a unique group of enzymes because, as the only genes in the proteome that, when translated by the rules of genetic coding, can then impose those rules, they compose a unique, reflexive interface between genes and gene products. This special relationship to the proteome lends considerable significance to the evolutionary phylogenetics of aaRS gene sequences [7,8], i.e. to how the aaRS came to be encoded. I will argue that by studying the ancestral coding of contemporary aaRS, we are led directly to a deeper understanding of how the genetic code might have arisen far more rapidly as a collaboration between ancestral proteins and RNAs than would ever have been possible in a world based entirely on a single polymer type [5,6].
There is consensus on several important aspects of aaRS structural and sequence-derived phylogenetics. Notably, they form two utterly distinct superfamilies that, on several levels are as distinct as possible from each other. Class I aaRS active sites all assume a Rossmann dinucleotide binding fold first observed in lactate dehydrogenase and flavodoxin [9] in which the active site forms at the interface between parallel β-strands and the amino termini of two helixes. In contrast, Class II aaRS active sites are formed from antiparallel β-strands.
These structural differences [10] motivated substantial effort to understand why and how nature would have divided the labor of tRNA aminoacylation in such a binary fashion [11–24]. Answers to these questions have emerged from supplementing phylogenetic analysis [25] with experimental deconstruction by protein engineering [26–31] and recapitulation [32,33] complemented by novel phylogenetic metrics [7,8].
Conclusions emanating from these studies change how we view the proteome and origin of genetic coding in important ways:
Class I and II aaRS appear to have originated from complementary coding sequences on opposite strands of the same bi-directional ancestral gene [26,34].
That gene complementarity has profound implications for the origin of genetic information, some of which had already been suggested by others [35–39].
The inversion symmetry of complementary coding strands has recognizable consequences for protein secondary and tertiary structures, and the active site construction of the resulting Class I and II enzymes [5,34], especially in light of the organization of the genetic code.
Complementary studies of the modular aaRS architectures of both synthetase classes [23,27] led to the discovery of how the organization of tRNA coding elements record how amino acids behave in water and in protein folding [40,41].
Whereas we can only begin to speculate on how such a gene emerged, it seems clear that it arose from a peptide•RNA partnership and not from an RNA World [5,42].
Thus, it now becomes possible to propose, in outline, a much more targeted program for studying how translation evolved.
B. The hypothesis of Rodin and Ohno [43]
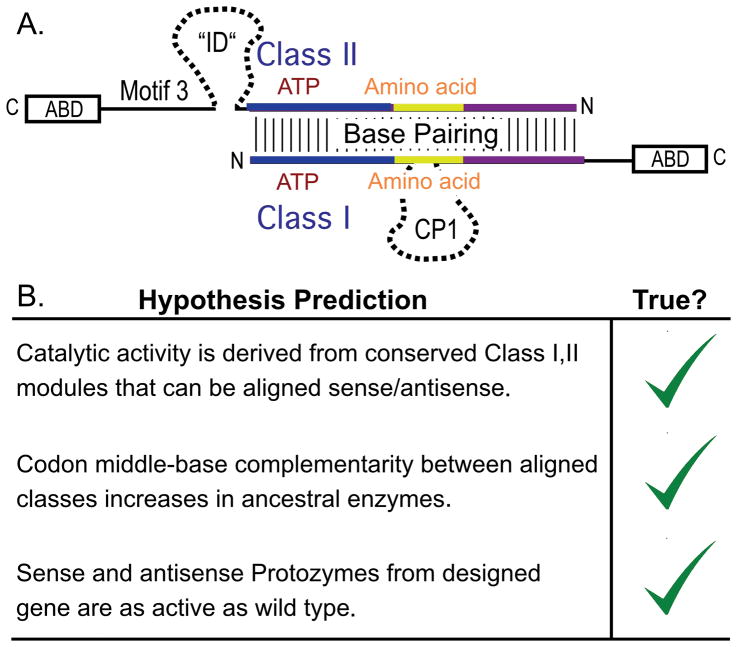
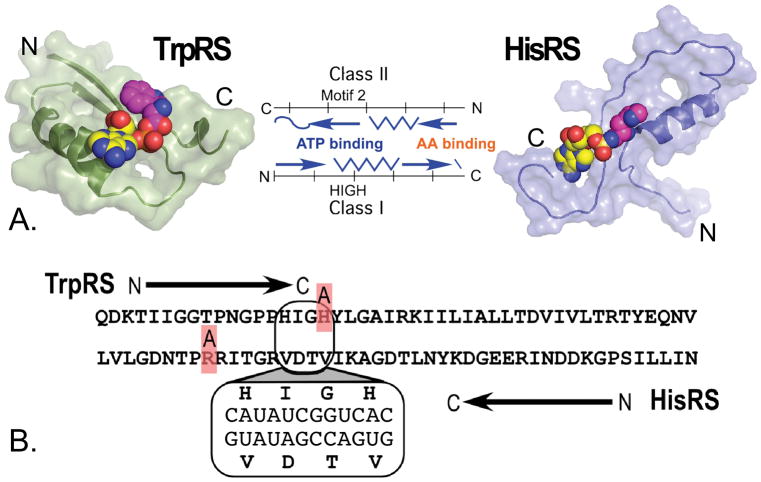
Shortly after the aaRS Class division became apparent [44,10,17,45,46] Rodin and Ohno published a remarkable hypothesis [43]. They used multi-family sequence alignments to establish consensus codons for the Class-defining motifs in the two superfamilies and found that codons for Class I PxxxxHIGH and KMSKS active-site catalytic motifs were almost exactly anticodons for Class II Motifs 2 and 1, respectively. That statistically significant, in-frame complementarity, illustrated in Fig. 1A, suggested that the contemporary aaRS superfamilies had at one time been coded by a single ancestral gene, one strand of which was transcribed and translated giving the ancestral Class I synthetase. Conversely, the opposite strand encoded the ancestral Class II synthetase.
Figure 1.
The hypothesis of Rodin and Ohno [43]. A. Schematic of the hypothesis. Genes for Class I and Class II aaRS are aligned in opposite directions as they would be oriented in an ancestral gene bearing the coding sequences on opposite strands. Vertical lines denote the extent of ancestral bi-directional coding, indicated by base-pairing of the coding sequences identified first in the Class-defining motifs in each superfamily. Large domains—the two anticodon-binding domains at the C-terminus of each Class and an insertion domain, ID in Class II, CP1 in Class I—are indicated by boxes and dashed lines, respectively. Substrate binding sites for the amino acid activation reaction, ATP and amino acid, are indicated. B. Summary of published evidence supporting the hypothesis, to be discussed in detail in this review.
The authors actually understated the statistical support for their case by not citing probabilities—10−8 – 10−18—of the observed alignments under the null hypothesis indicated by their jumble-testing z-scores. Perhaps for this reason, the hypothesis remained more or less dormant for almost a decade before it was revived by Carter & Duax [47,48]. Subsequent work has substantially strengthened the experimental and bioinformatic support for the hypothesis by articulating and testing predictions that it makes (Fig. 1B). Direct support for the hypothesis, discussed at length in this review, is made more relevant by related work on the origin of translation [5,49–54] and the genetic code [40,41,55].
C. The origins of symbolic interpretation and coding
Biological nucleic acid sequences represent an exquisite repository of information relevant to managing stimuli from the world at large. For our purposes, it is useful to distinguish between two types of information stored in nucleic acids. Information about the chemical environment of biology defines how amino acids behave in water; gene sequences exploit that information by configuring amino acid sequences capable of folding into functional proteins.
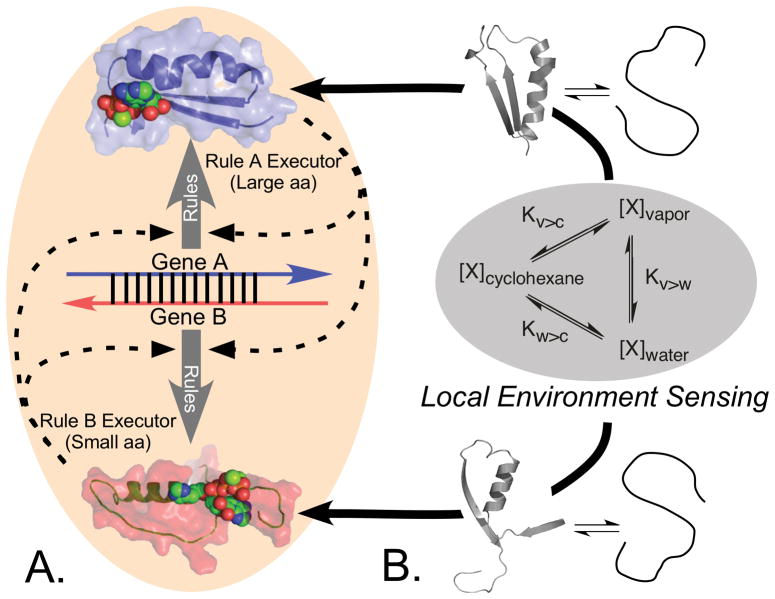
The information in genes furnishes the blueprints for assembling proteins via the ribosomal read-write apparatus. Genes constitute a set of programs, written in the language of the genetic code, and expressed as a sequence of codons, or symbols consisting of three consecutive nucleotide bases, each with a specific meaning—start, a particular amino acid belongs here, stop. Equally important is the translation table embedded in transfer RNA. This second type of information specifies the conversion of the symbolic information in codons into specific amino acids. It has recently become apparent that this conversion corresponds closely to the phase transfer equilibria that enable translated gene sequences to fold and function. It represents the programming language in which genes are written and self-organization has embedded it efficiently and robustly into tRNA base sequences, primarily in the acceptor stem and anticodon.
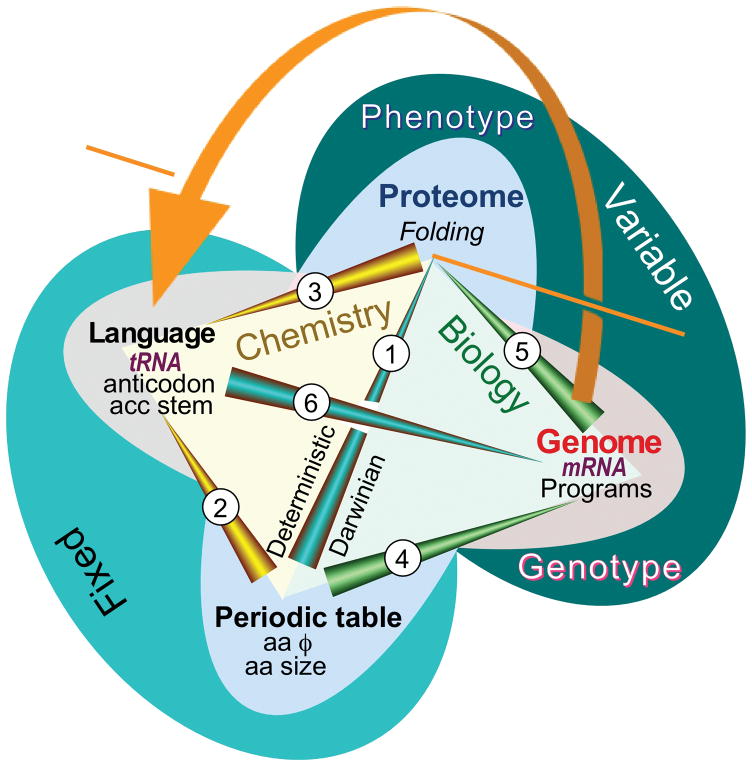
The aaRS connect codons, hence messages in mRNA to amino acids via the translation table in the genetic code, each synthetase performing specific tRNA aminoacylation to enforce the rule specifying that a particular codon means that a particular amino acid is to be inserted whenever the anticodon of its cognate tRNA matches a codon in the message. As the synthetases are themselves made according to specific mRNA sequences, their connection to the genetic code is deeply reflexive or self-referential: once translated, they themselves become sensitive to the impact that water has on their constituent amino acids and fold into active conformations. Those folded conformations subsequently execute the symbolic rules in the genetic coding table to make themselves and all other proteins (Fig. 2; [40]).
Figure 2.
Aminoacyl-tRNA synthetases and their cognate tRNAs furnish the reflexive elements necessary to translate the genetic code (orange arrow). The code connects their gene sequences, via their folded structures, to the enzymes that can enforce the coding rules in the codon table. Network analysis of the Central Dogma of Molecular Biology accommodates new evidence relating both tRNA identity elements and protein folding directly to the free energies of phase transfer equilibria of amino acid side chains [40,41,55]. Two of the nodes of a tetrahedron—physical properties of amino acids and the codon assignment table—reside in the realm of chemistry. They are “fixed” because they obey chemical equilibria. The other two nodes—gene sequences and protein folding—are dynamic processes that transcend chemistry because they are variable and form the basis for the evolution of diversity through self-organization and natural selection. The network connects RNA and Protein worlds via six edges: 1. Protein folding depends on both amino acid polarity and size [55], properties that play a role analogous to those of elements in chemistry, in that the amino acids form a kind of “periodic table” on which the folding of proteins is based. 2. tRNA bases encode amino acid size and polarity separately [41]. The acceptor stem codes for amino acid sizes; the bases of the anticodon code for amino acid polarities. Amino acid properties therefore dictate how tRNA bases are recognized by aminoacyl-tRNA synthetases. 3. tRNA codes are related to protein folding. Together, statements (1) and (2) imply that for the aminoacyl-tRNA synthetases, the code (and mRNA sequences) must define folded structures that bind specifically to particular tRNAs in order to read the language of genetics. The bi-directionality of this arrow illustrates a somewhat deeper self-referential element than those identified in molecular biology by Hofstadter as generators of complexity according to Gödel’s incompleteness theorem [179]. 4. Gene sequences (mRNA) are analogous to computer programs. Genetic instructions assemble amino acids according to their physical properties in ways that, when translated according to the programming language in tRNA (in 5), yield functional proteins (enzymes, motors, switches, regulators). 5. mRNA sequences (i.e., the genotype) determine amino acid sequences in proteins, and hence how amino acid sizes and polarities are exploited to produce different folded proteins. The spontaneous folding of amino acid sequences gives rise to functions (i.e., the phenotype) that ultimately determine whether or not a particular sequence survives natural selection. Changes accumulated in gene sequences result from selection acting on the phenotype. (4) and (5) localize how selection incorporates information about amino acid behavior into gene sequences, hence depict the evolutionary dimension in biology. 6. The evolution of mRNA sequences only makes sense in the context of the translation table (or programming language) established by the genetic code. (Adapted, with permission, from Carter, CW, Jr & Wolfenden, R. (2016) RNA Biology 13:145–151.)
At the nucleic acid level, the molecular nature of this information, and how it is preserved from one generation to the next, are well understood in terms of base-pairing, as are the general structural mechanisms by which this base sequence information is read out by transcription, and converted first to protein sequences by the ribosome during translation and then to folded, active enzymes. Principles of protein folding are also beginning to be understood, at least in outline [56–58]. Key to understanding the origin and evolution of each of these mechanisms is the element of “interpretation”—rephrasing the encoded information into a more flexible form capable of a greatly extended range of functionalities.
How the genetic code became embedded in tRNA sequences and how mRNA sequences originated, however, have been essentially blank pages. High probability synthetase•tRNA complexes were essential to launching translation. The probability of implementing molecular recognition and interpretation via self-organization [59,60,51,61,62] and natural selection [63] decreases sharply the more sophisticated the system. Thus, it seems likely that translation began with smaller, less specific complexes, hence a simpler, less precise, and probably redundant alphabet. The recent experimental work reviewed here points clearly to molecular models with just those properties. These and other arguments [5,6]. imply that genetic coding arose in a flexible, rudimentary implementation that later underwent successive refinements that completed the code [5].
Given that the aaRS are probably the first and only gene products (ie., of the second type of information) with the ability to interpret the first type of information (ie., the genetic code translation table), it should come as no surprise that their molecular phylogenies contain potentially useful information concerning how they arose and began translating genetic messages. Retracing evolution is a unique subset of reconstructing the past, i.e. of history. It can be argued that the effort is fruitless as one cannot run the tape in reverse, that important and relevant witnesses are all extinct, and in particular that there is no way to test hypotheses. Our work [64,40,65,41,55,26,42,32,34,27,7,28,33,29,8,30,36,31,48], and that of others [25,66,52–54,67], tends to rebut this objection. In reality, molecular phylogenies of the Class I and II aaRS have proven to be a rich source of unexpected insights into how translation became possible and robust tools now enable us to investigate, and even recapitulate key events from the past history of life.
II. Evidence for Bi-directional Coding Ancestry: Molecular Phylogenies, Urzymes, and Protozymes
The broader evidence now supporting the hypothesis of Rodin and Ohno [43] began with analysis of superpositions of the three-dimensional structures of Class I and II aaRS, as illustrated in Fig. 1 and described in §II.A. §II.B reviews the experimental characterization of the parallel catalytic activities and amino acid specificities of the deconstructed hierarchies from both classes. Problems raised by experimental recapitulation of putative evolutionary events connecting the ancestral forms to the contemporary enzymes are discussed in §II.C. A new distance metric for phylogenetic analysis of protein superfamilies related by bi-directional coding ancestry is reviewed in §II.D, together with its possible use in identifying how synthetases for the 20 canonical amino acids may have diversified from a single ancestral gene.
A. Protein engineering and experimental deconstruction
The overall strategy of these studies has been to deconstruct Class I and II aaRS into genes for their component modules, use enzyme kinetics to characterize their catalytic activities and specificities, and validate their authenticity. Recapitulation of putative evolutionary intermediates by partial reconstruction also has been carried out, although to a lesser extent, as described in §II.C.
Deconstruction
Genes coding for intermediate modules were made using molecular biological techniques. For Class II aaRS in which the active site is formed by a continuous, uninterrupted coding sequence, deconstruction can be accomplished using PCR amplification of the desired region [29]. For Class I aaRS, however, the active site—and Urzyme—are discontinuous, requiring more aggressive protein engineering [30,68]. Two aspects of the fusion and solubilization of the Class I Urzymes required amino acid sequence modification: (i) the intervening insertion element had to be removed and the two ends fused together, and (ii) an extensive surface area of nonpolar side chains, exposed by the removal of entire domains, needed to be modified to enhance solubility. In constructing Urzymes for TrpRS and LeuRS, both operations were accomplished using the Design module in the Rosetta program [69].
Urzymes
Multiple sequence and especially multiple structure alignments furnish the basic tools for constructing molecular phylogenies [25] (Fig. 3). Superimposing three-dimensional structures of proteins within the same superfamily reveals that certain modules are shared by all family members, whereas others differ distinctly from member to member. The most conserved modules generally contain the active sites, and for that reason alone are likely candidates for evolutionary intermediates. For both aaRS classes, modules shared by all ten superfamily members contain essentially their intact active sites built from ~130 residues [31,48]. These modules have been expressed independently of the rest of the contemporary gene from two Class I and one Class II aaRS and shown to exhibit ~60% of the transition state stabilization of the full-length enzymes [29–31]. Their extensive conservation and enzymatic activities earned them the descriptor “Urzyme” from the German prefix Ur = primitive, authentic, original plus enzyme.
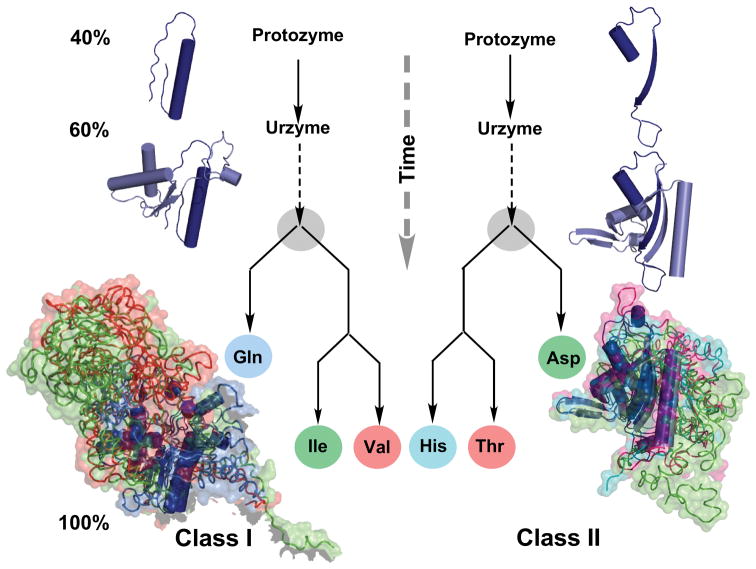
Figure 3.
Inferring protein family trees from molecular anatomies. Structures of three class I and class II aaRS have been rotated into a common orientation using their atomic coordinates and colored differently. α-Helical secondary structures are drawn as cylinders; extended β-structures as ribbons with arrows indicating their direction. Larger, more differentiated structures are drawn as noodles, surrounded by their surfaces. Differences in the recent structures at the bottom are highlighted by modules of one color that are absent in other structures. Such differences can be quantified and used to construct the genealogies in the center. Modules that are most similar in all three are colored dark blue and are inferred to be present in the common ancestor. Circles represent essentially modern aaRS. The three structures in each aaRS class are labeled with their three-letter abbreviations. There is consensus that they were present together with twelve other aaRS in the last universal common ancestor (LUCA) of all living organisms [180,181]. Novel results described here are the construction, expression, and experimental testing of ancestral forms called urzymes and protozymes, which are found, essentially without variation in all contemporary species and which retain substantial fractions—60% and 40% respectively—of the catalytic activity of the contemporary enzymes. The similarity between the class I and II genealogies is evidence that the two families evolved coordinately. (Courtesy of Carter Natural History CW, Jr. (2016) Natural History 125:28–33.)
Protozymes
Mildvan published a series of studies in which he excerpted the ATP binding sites of three different P-loop ATPases—F1 ATPase [70,71], DNA polymerase [72], and adenylate kinase [73,74]—and demonstrated that they retained ligand-dependent structures similar to that observed in the full-length proteins. All three ATP binding sites consist of ~50 residue β-α-β secondary structures with a glycine-rich loop between the first strand and helix, and appear homologous in these respects to the Class I aaRS ATP binding sites. That precedent motivated further deconstruction of the Class I and II Synthetase Uryzmes, both of which contain ATP binding sites of approximately the length—46 residues—studied by Mildvan. Expression and fluorescence titration of ATP by these 46-mers established that they, too, bind ATP tightly, motivating investigation of their possible catalytic properties. ATP binding sites from both Class I and II aaRS accelerated amino acid activation by 106-fold [26], and led to their designation as “protozymes” from “proto” = first.
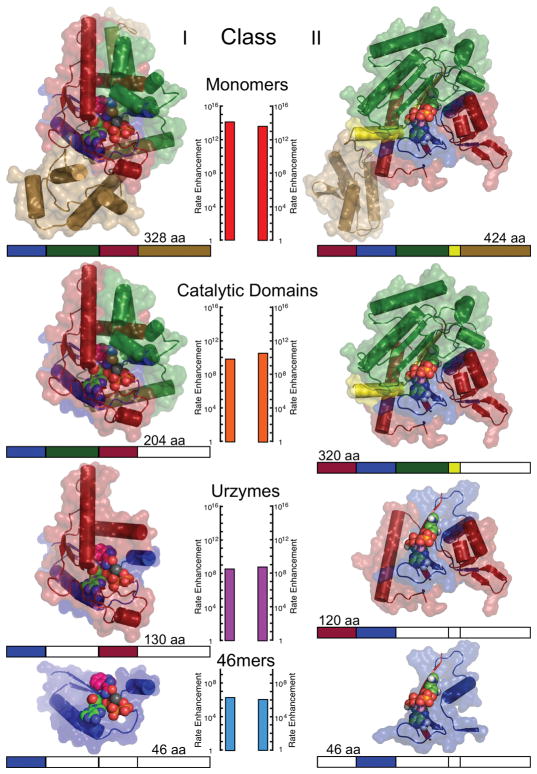
The hierarchy—monomer>catalytic domain>Urzyme>protozyme (Fig. 4)—illustrates the parallel evolution of both Class I and Class II aaRS providing details abstracted in Fig. 3. Of particular interest are the following:
Figure 4.
Deconstruction of Class I tryptophanyl (PDB code 1MAU)- and II histidyl (PDB code 2EL9)-tRNA synthetases into successively smaller fragments that retain catalytic activity (1, 7–10, 43). Graphics for smaller constructs are derived from coordinates of the full-length enzymes. Colored bars below each structure denote the modules contained within each structure; white segments are deleted. The number of amino acids (aa) in each construct is noted. Measured catalytic rate enhancements for 32PPi exchange, relative to the uncatalyzed second-order rate (kcat/Km)/knon are plotted on vertical scales aligned in the center of the figure and are colored from blue (slower) to red (faster). (This research was originally published in the Journal of Biological Chemistry Martinez, L. et. al., (2015) Journal of Biological Chemistry 290:19710–19725 ©American Society of Biochemistry and Molecular Biology)
Red and blue modules are interrupted by an insertion (connecting peptide 1 CP1 [75]) in the Class I Uryzme but continuous in the Class II Urzyme.
The protozyme module (blue) occurs at the amino terminus of the Class I and at the carboxy terminus of Class II aaRS Urzymes.
Transition-state stabilization free energies for amino acid activation assayed by PPi exchange for each catalyst, ΔGkcat/KM = −RTln(kcat/KM), are approximately linearly related to its mass [26].
Catalytic activities of Class I [30,68]and II [28,29] Urzymes were the first observations to substantively validate predictions implied by Rodin and Ohno for the bi-directional genetic coding of Class I and II aaRS. The third observation establishes a crucial pre-requisite for the evolution of catalytic activity in general: insofar as catalysis is required to synchronize the rates of chemical reactions in the cell, it is essential that different enzyme families across the proteome evolve so as to preserve parallel increases in rate enhancement.
Characterization
Overexpressing Urzymes from both Classes leads to their accumulation in inclusion bodies. Washed inclusion bodies contain >50% Urzyme in such cases, and therefore represent a significant purification. Inclusion bodies solubilized in 6 M guanidinium hydrochloride can be renatured by size exclusion chromatography on superdex 75, which also yields essentially pure Urzyme. Active-site titration [76,77] shows that between 35–70% of the molecules in various preparations contribute to the observed activity seen in pyrophosphate exchange assays [77]. TrpRS and HisRS Urzymes accelerate the rates of amino acid activation (assayed by pyrophosphate exchange) and tRNA aminoacylation by 109-fold and 106-fold, respectively [28]. These values are consistent with measurements of the uncatalyzed rates for the two reactions, as spontaneous amino acid activation [78] is ~1000-fold slower than are either spontaneous acylation [79] and peptide bond formation from activated amino acids [80,81].
As protozymes isolated from the two aaRS Classes have only ~40% of the mass of Urzymes, they are substantially weaker catalysts, activating cognate amino acids 106 times faster than the uncatalyzed rate. PPi exchange assays were incubated for 14 days and assayed at intervals of several days [26]. The specificities of amino acid activation by wild-type and bi-directionally coded protozymes, and their possible acylation activities have yet to be determined.
Validation
Establishing the authenticity of the catalytic activities observed for the aaRS Urzymes and protozymes is obviously of great importance, and is a matter to which considerable attention has been paid [34,27–31]. They are much weaker catalysts than full length enzymes, and consequently, their activities can much more readily be attributed to very small amounts of various kinds of contaminating enzymes, including, of course, the full-length native homologs present in all cell extracts. In addition to the absence of activity in conventional controls carried out using extracts prepared from cells containing empty cloning vectors, authenticity was established by four controls:
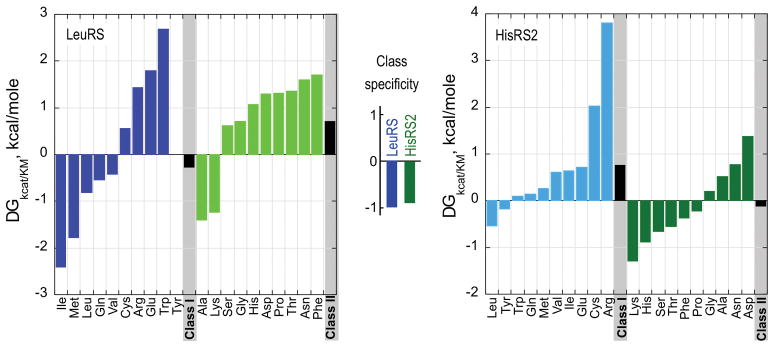
Steady-state kinetic experiments show that Urzyme and protozyme activities saturate at amino acid concentrations several orders of magnitude higher than is required to saturate the full-length enzymes. This argument is strengthened by the complete specificity spectra determined for the Class I LeuRS and Class II HisRS Urzymes (Fig. 5; [42,34]).
Cryptic catalytic activity is released when Urzymes expressed as fusion proteins with maltose-binding protein are treated with TEV protease.
Active-site mutations and modular variants containing minor additional mass at the N- and C-termini alter the measured activity.
Active-site titration confirms that a major fraction of molecules contribute to the observed activity (aaRS Urzymes only; it is unclear that the protozymes would exhibit a pre-steady state burst, which is a requisite for active-site titration).
Figure 5.
Amino acid specificity spectra of Class I LeuRS and Class II HisRS2 Urzymes.
The mean net free energy for specificity of Class I and II Urzymes for homologous vs. heterologous substrates, i.e., ΔG(kcat/KM)ref – ΔG(kcat/KM)amino acid (i), where ref is the cognate amino acid, is approximately 1 kcal/mole for both Class I and II Urzymes (center). Class I amino acids are colored blue; Class II amino acids are colored green. Bold colors denote substrates from the homologous Class; pastel colors denote heterologous substrates. (From Carter, CW Jr., (2015) MDPI Life 5: 294–320).
All these results implicate the actual genetic construct in the observed activity, and contaminating activities cannot account for either (ii), (iii), or (iv).
B. Class I and II aaRS deconstructions exhibit parallel catalytic hierarchies
Catalytic rate enhancements correlate with catalyst mass
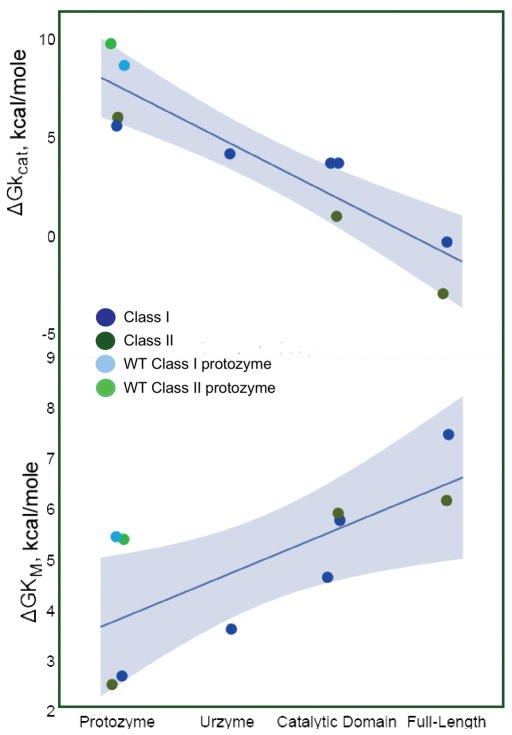
Deconstructions (Fig. 4) reveal surprisingly consistent increases in transition-state stabilization with additional masses in the ascending hierarchies [42,34]. Catalyst masses range by 70-fold from ~6.5 KD to 450 KD, and the constructs derived from each Class are distributed differently with respect to size. Class I deconstructions are a “low resolution” map because they include two modular hybrids—catalytic domain and Urzyme plus anticodon-binding domain—between the Urzyme and the full length monomers. The Class II constructs, on the other hands, include high resolution divisions—increments of 6, 20, and 26 residues—at approximately the 126-residue size of the Urzyme. Across this entire range of deconstructions, transition-state stabilization energies increase linearly with the number of residues [26]. Moreover, the slopes for each Class are the same within 5%.
Class I, II constructs at each stage have the same catalytic proficiencies
A second remarkable result of the aaRS deconstruction is that the linear relationships between transition-state stabilization free energy and the number of residues also have the same intercepts. This implies strongly that throughout the evolutionary history of the two synthetase Classes, they retained comparable catalytic proficiencies [26]. The importance of this observation is that the synthetase superfamilies form a tightly interdependent autocatalytic set coupled by the fact that each is required to operate with approximately the same throughput of aminoacylated tRNAs for translation of all amino acids within the current genetic alphabet [5]. Their enzymatic activities must, therefore, have remained quite comparable for all relevant amino acids over the duration of the synthetase superfamily growth from short peptides to long polypeptides. It was not obvious, however, that experiments would confirm that expectation. Nevertheless, aaRSs from both Classes appear to have been capable of parallel increases in both size and catalytic proficiency, consistent with continuously providing comparable quantities of aminoacyl-tRNAs for all amino acids throughout the evolutionary tuning of the genetic code.
Class I and II Urzymes are promiscuous catalysts that nonetheless have comparable amino acid specificity
Essentially complete amino acid specificity spectra have been determined for amino acid activation by the Class I LeuRS and Class II HisRS Urzymes (Fig. 5) [42,34]. Remarkably, the two catalysts retain a significant preference for the class of amino acid substrates for which their parent enzymes were specific. The LeuRS Urzyme prefers not to activate Class II amino acids; the HisRS2 Urzyme prefers not to activate Class I amino acids. The degree of specificity, evaluated as the free energy of the specificity ratio, ΔGkcat/KM(I/II) and ΔGkcat/KM(II/I), are ~ − 1 kcal/mole for both Urzymes. This value is roughly 20% of that for the full-length enzymes. It means that given equimolar concentrations of all 20 amino acids, the Class I Urzyme will activate an amino acid from the wrong class roughly one time in 5, whereas a native aaRS will typically active an incorrect amino acid roughly one time in 5000. Thus, aaRS Urzymes are promiscuous with respect to amino acid recognition, but retain the Class preferences of the full-length enzymes from which they were derived for amino acids within their own class.
As noted below, the problem of evolving high amino acid specificity is more subtle than might appear from the initial studies in Fig. 5. An important possibility is that specificities were enhanced in the presence of cognate tRNAs, as is true for several contemporary aaRS [82–85]. Work is in progress to characterize tRNA specificity in a similar fashion, and to determine whether or not the amino acid specificity spectra (Fig. 5) improve in the presence of cognate tRNAs.
Wild-type and bi-directional protozyme gene products from both Classes have the same catalytic proficiencies
§II.D discusses in greater detail the extent to which experimental and bioinformatics results have confirmed the hypothesis of Rodin and Ohno that the original ancestral genes for Class I and II aaRS were fully complementary. It is worthwhile noting here that wild type Class I and II protozymes were excerpted directly from full-length TrpRS and HisRS genes. Although those coding sequences retain a strong trace of their bi-directional coding ancestry, they are distinctly not complementary. To test the prediction that a fully complementary protozyme gene could be achieved, the computer design program, Rosetta, already used extensively in the re-design of Class I Urzymes, was adapted to impose coding complementarity on the two protozyme genes, resulting in a single gene with two different functional translation products, one from each strand [26].
Analysis of the peptides coded by the resulting bi-directional gene showed that the four gene products—Class I and II; designed and wild type—have nearly the same catalytic proficiency, ΔGkcat/KM = +3.5±0.8 kcal/mole. The amino acid sequences of the designed protozymes are quite different from the WT sequences. This agreement therefore suggests that the catalytic activities of the Class I and II protozymes may be consistent with a very large number of different sequences that share only simple patterns based on a reduced alphabet of fewer amino acids, consistent with their possible emergence at a time when the amino acid alphabet was both smaller and less faithfully implemented.
Designed protozymes have high turnover, low specificity
The steady-state kinetic parameters for WT and designed protozymes revealed yet another remarkable comparison. Although the overall second-order rate constants, ΔGkcat/KM, are very nearly the same, their similar values arise from quite different values for the turnover number and amino acid substrate affinity. The WT protozymes, perhaps because they were excerpted from the full length proteins, retain higher ground-state substrate affinities but have lower turnover numbers, whereas the designed Protozymes have higher turnover numbers and weaker ground-state affinities [26]. The differences in both parameters are about 100-fold, leaving their kcat/KM ratios unchanged.
Without intending to do so, by enforcing genetic complementarity the design process also enhanced the turnover number while weakening amino acid affinity (Fig. 6). Specific binding of cognate, versus non-cognate amino acids cannot be improved without increasing the binding affinity of the cognate complex, so increased ground state amino acid affinities are a prerequisite for enhanced discrimination between competing substrates. Thus, deconstructions of both aaRS Classes exhibit parallel enhancements that improved fitness by increasing both catalysis and specificity. Notably, the higher turnover number and lower amino acid affinity of the designed, bi-directionally coded protozymes match properties expected for a emerging rudimentary coding apparatus.
Figure 6.
Parallel sequential improvements in catalytic rate enhancement (top) and ground-state amino acid affinity (bottom) associated with modular enhancements of Class I (blue) and Class II (green) aminoacyl-tRNA synthetase evolution. Increased amino acid specificity requires higher affinity binding of cognate amino acids, to differentially release non-cognate complexes. Thus, the behavior of the ground-state amino acid affinity is a surrogate for specificity. Both catalytic proficiency and specific recognition of cognate amino acids therefore improve with more sophisticated modularity. Pastel colors show that imposing bi-directional coding on the protozymes increases both kcat and KM proportionately.
C. Recapitulation
Modular engineering of TrpRS
One of the best ways to validate and utilize knowledge gained from the reconstruction of ancestral forms is to recapitulate putative evolutionary steps by reconstructing and testing intermediates [86–88]. The deconstruction of the Class I and II aaRS has afforded such opportunities [33,29]. Those investigations cast new light on synthetase function and evolution.
Two putative TrpRS constructs intermediate between the Urzyme and full-length enzyme involved the re-insertion of the CP1 fragment to restore the catalytic domain and the covalent joining of the anticodon-binding domain to the Urzyme [33]. Comparison of these modular variants showed that, although both intermediate species exhibited modest increases in catalytic proficiency, neither was any better than the Urzyme, either in aminoacylation or in discriminating between cognate tryptophan and non-cognate tyrosine [33]. There are two notable interpretations of this surprising result. First, as neither intermediate construct would have sufficiently increased fitness to be selected, it suggests that the apparently separate evolutionary enhancements must have occurred coordinately, either because one of the two had already begun to function in trans, or because one or the other, or both modules could “grow” by smaller modular additions that did endow enhanced fitness [33]. Second, the modular thermodynamic cycle involving full-length TrpRS, the two distinct intermediates [33], and the Urzyme allowed measurement of a ΔΔG‡ ~ − 5 kcal/mole coupling energy between the CP1 and anticodon-binding domains in the transition state of the amino acid activation reaction by full-length TrpRS [33], shedding new light on the general problem of intramolecular signaling or allostery [89–91].
Mechanistic studies on intact TrpRS had previously identified a profound intramolecular coupling, ΔΔG‡ ~ − 5 kcal/mole necessary for catalytic assist by the active-site Mg2+ ion [92] and achieving full catalytic proficiency by the full-length enzyme [89,32,93,92]. A five-way coupling interaction, ΔΔG‡ ~ − 5kcal/mole, was also measured between four residues in an allosteric switching region 20 Å from the active-site metal that mediates the shear involved in domain movement during catalysis [94]. A related study [32] confirmed that the same coupling energy was used in the transition state to enforce the specific selection of cognate tryptophan vs non-cognate tyrosine. Thus, the modular thermodynamic cycle provided a key link connecting the long-range coupling observed previously directly to the domain movement: the four switching side chains (I4, F26, Y33, and F37), the Mg2+ ion, and both domains all move coordinately in the transition state [89,95].
Modular engineering of HisRS
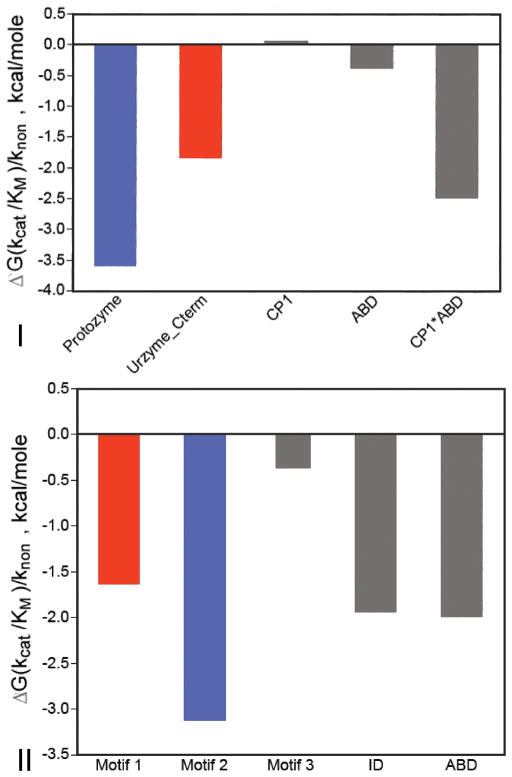
Three conserved motifs are recognized in Class II aaRS: Motif 1 and Motif 2 compose the HisRS Urzyme. The third, Motif 3, however, lies well outside the Urzyme. It is separated by a long and variable insertion domain, C-terminal to the Urzyme, much as the long and variable CP1 insertion interrupts the Class I Urzyme. Exploratory modular engineering of interactions in the Class II HisRS yielded several intriguing observations [29]. (i) Motif 3 could be fused together with the HisRS Urzyme to produce a module whose catalytic activity is intermediate between that of the Urzyme and that of Ncat, the HisRS catalytic domain containing both Motif 3 and the insertion domain [96]. (ii) Catalytic proficiency of the Motif 3-supplemented HisRS Urzyme is further enhanced by adding six additional residues N-terminal to the Urzyme [29]. (iii) The six-residue N-terminal fragment functions synergistically with Motif 3. Effects of the five modules estimated by regression methods from all of the measurements (Fig. 7) distinctly resemble those evaluated on the basis of more thorough investigations of Class I constructs.
Figure 7.
Corresponding modules make similar relative contributions to catalysis by Class I and II aaRS. The Protozymes are colored blue; the remainder of the Urzymes red; and additional modules, including the insertion domains and anticodon-binding domains grey. Differences between the two Classes include the fact that Class II Motif 3 has no corresponding module in Class I aaRS, and the insertion and anticodon-binding domains have not been examined separately in Class II, so their synergistic interaction, which is quite large in Class I aaRS, cannot be estimated.
D. Middle codon-base pairing: a new phylogenetic distance metric
Bi-directional genetic coding left a detectable trace in contemporary sequences
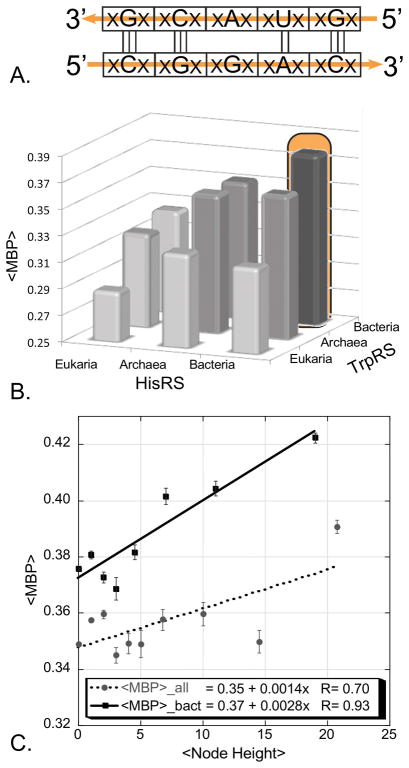
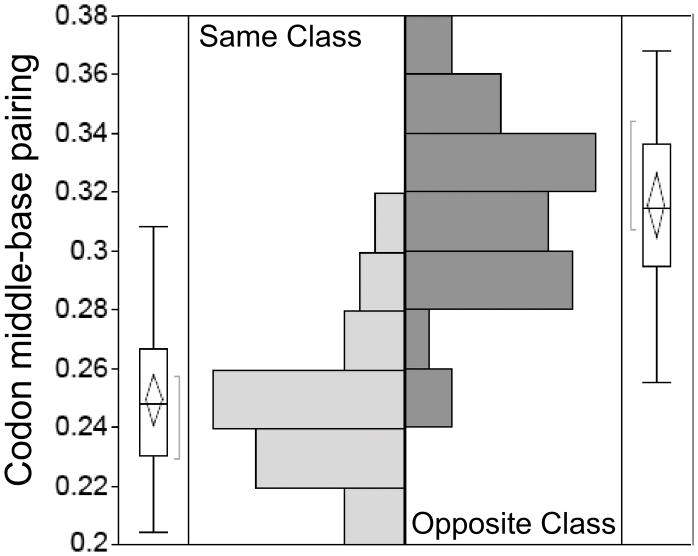
Sense/antisense alignment of coding sequences from different protein families introduced a new, phylogenetic distance metric—the percentage of middle codon bases that are complementary in all-by-all in-frame bi-directional alignments of multiple sequence alignments from the two families. As an example, aligning the TrpRS Class I Uryzme against the HisRS Motif 2 [31] revealed that the region of quite extensive codon-anticodon complementarity identified by Rodin and Ohno could be extended to include ~75% of both Urzyme sequences, provided that the first and third codon bases were excluded. Outside regions of very high conservation as found in the Class-defining signatures of the aaRS, a transient ancestral use of dual strand coding, followed by an extended period of adaptive radiation would rapidly degrade the complementarity of the two strands. The highly conservative nature of the genetic code, together with wobble property of the third codon base [97] mean that loss in the middle-base pairing occurs much more slowly as sequences diverge than that in the first codon bases on each strand, each of which is opposite a wobble base on the opposite strand (Fig. 8A). The trace of bi-directional coding ancestry can thus be recovered by structurally-informed middle codon-base alignments of sufficient numbers of contemporary sequences (Fig. 8B) [7]. To wit, if the number of sequences aligned is sufficiently high (~104 comparisons in ref. [31]), the standard error of the mean is reduced to a tiny fraction of the differences between the pairing frequencies of Class I vs Class II alignments and those (0.25) expected under the null hypothesis that one base in four would be complementary.
Figure 8.
Codon middle-base pairing (<MBP>) furnishes a new phylogenetic distance metric. A. Transient bi-directional coding is rapidly lost after the constraint is released. First and third bases lose complementarity much faster than the middle bases, which retain residual base-pairing into contemporary sequences. B. Comparison of middle codon-base pairing in all-by-all alignments of “Urgene” sequences excerpted from ~200 contemporary HisRS and TrpRS sequences [7]. C. Node-dependence of middle codon-base pairing in antiparallel alignments of middle bases from ancestral sequences reconstructed independently for the TrpRS and HisRS Urzyme multiple sequence alignments. Solid line is for bacterial sequences, dashed line is for all bacterial, archaeal, and eukaryotic sequences. (B and C adapted with permission from the Society for Molecular Biology and Evolution. Chandrasekaran, SN et. al. (2013) Mol. Biol. Evol. 30: 1588–1604.)
Elevated codon middle-base pairing in multiple antiparallel alignments between different Class I and II aaRS coding sequences occurs generally throughout all aaRS superfamilies
The significance of the published study of middle codon-base pairing [7] raises a potential question because the statistics were accumulated for alignments of a Class IC (TrpRS) with a Class IIA (HisRS) synthetase. Neither of these synthetases was likely to have been among the earliest to appear. To establish the significance of the middle codon-base pairing distance metric, we therefore extended this analysis to include eleven aaRS, balancing the three subclasses by including six from Class I (TrpRS, TyrRS, LeuRS, IleRS, GluRS, GlnRS) and five from Class II (HisRS, ProRS, AspRS, AsnRS, PheRS; N. Chandrasekaran, personal communication). The alignments included 64 amino acids surrounding the PxxxxHIGH and KMSKS sequences in Class I aaRS and the Motif 1 and 2 sequences in Class II aaRS in [7] to enhance confidence. The trace of ancestral bi-directional coding remains significant.
This extended database samples multiple comparisons between all subclasses within each Class, and hence include pairs of aaRS that presumeably appeared at different times along the evolution of the code. The statistical structure of this new database is shown in Fig. 9. The bi-directional alignments add an average of ~0.07 ± 0.007 to the fraction of codon middle-base pairing over those within the same aaRS Class. The difference between within- and between-classes accounts for ~60% of the variance in observed pairing (R2 = 0.60), and the Student t-test probability for a ratio of the slope to its standard error as large as 10 is ~10−14.
Figure 9.
Extended analysis of mean codon middle-base pairing. Results from [7] have been updated to include alignments of ~200 sequences from each of eleven contemporary synthetases (Asp, Asn, Glu, Gln, His, Ile, Leu, Pro, Phe, Trp, Tyr). Histograms are shown for the fraction of base pairing between middle codon bases for antisense alignments from the same and opposite aaRS Classes, as indicated. The difference between the two mean values indicated by horizontal lines in the logos outside the histograms is 0.067, which is ~14 times the standard error. Further statistics of the comparison are discussed in the text and shown in Fig. 10.
Ancestral sequence reconstruction extends the phylogenetic evidence for bi-directional coding significantly backward in time
Ancestral gene reconstruction [98,99] has become broadly used to resurrect ancestral enzymes [100] and signaling proteins [101–103,87,104,88,105]. Given the evidence for significant residual codon middle-base pairing in contemporary Class I and Class II sense/antisense alignments (Fig. 8B), it was of interest to extend the technique to the quantitative comparison of distinct gene families whose evolutionary descent might have been tightly coupled by bi-directional genetic coding at the origins of translation. That prediction led to the expectation that reconstructed node sequences of both superfamilies might exhibit increased codon middle-base pairing as reconstructed nodes from each family are aligned in opposite directions. This test is distinct from the construction of phylogenetic trees from multiple sequence alignments of related proteins, because it compares separate reconstructions of distinct families carried out independently and aligned only after the nodes have been reconstructed. The resulting appearance of increased codon middle-base pairing (Fig. 8C) is therefore a significant, orthogonal verification of the Rodin-Ohno hypothesis [7].
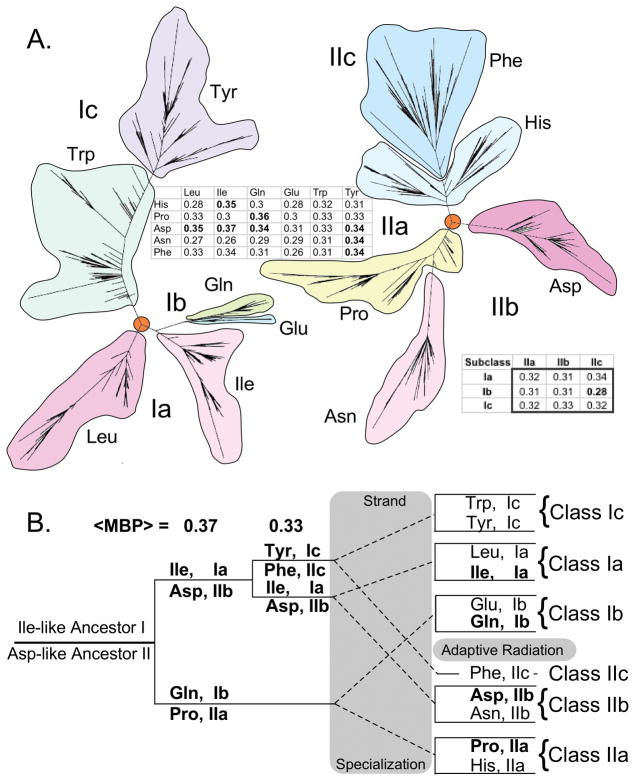
Codon middle-base pairing may contain evidence for very early stages of genetic coding
The breadth of the histograms in Fig. 9 and consensus subdivisions of the two aaRS Classes into parallel subclasses, one large and two small, suggest that further examination of the middle-base pairing metric may eventually provide clues about the order in which pairs of aaRS speciated, and hence the order in which amino acids appeared in coding relationships. We constructed putative phylogenetic trees from the aligned amino acid sequences of eleven aaRS (six Class I and five Class II; Fig. 10A) and from the middle base-pairing distance metric (Fig. 10B) to illustrate this possibility. Significantly, there is only one significantly lower middle codon-base pairing metric among the all-by-all comparison of the subclasses—subclasses Ib and IIc appear to be more distantly related than all of the other subclasses. Thus, the distance metric implies comparable distances between all aaRS subclasses of each class to those of the other. Although based on partial data, this analysis is nevertheless interesting because it suggests ancestral genes in which the two principal subclasses are swapped (Fig. 10B): strands of the presumptive ancestral gene encoded ancestral Class Ia Ile-like and Class IIb Asp-like protozymes. Similarly, the next most prominent middle-base pairing metric relates sequences of Class IIa ProRS those of Class Ib GlnRS. Further work in this direction is in progress, and will require developing improved analytical tools for using the new distance metric, along with ways to deal with the ancestral sequence reconstructions built from amino acid alphabets of decreasing size [49,106,107].
Figure 10.
Extended analysis of the middle codon-base pairing metric confirms previous statistical evidence for ancestral bi-directional coding. Approximately 200 sequences, broadly distributed among the three canonical domains of life were assembled for 64 amino acid residues from 11 of the 20 canonical aminoacyl-tRNA synthetases. The 64 residues included the 46 residues of the protozymes together with 18 residues of the KMSKS and Motif I loops from the respective aaRS classes. Tables embedded in the figure show the mean middle codon-base pairing metrics for the all-by-all comparison of the 11 aaRS, and the corresponding average values for the canonical subclasses. A. Independent phylogenetic trees drawn from the complete 192-base sequences of each class. B. A putative tree drawn according to distances from the middle codon-base pairing metric for a 64-base subset of the sequences in A.
These data suggest that we now potentially have the tools to address directly the question of which stepwise bifurcations were actually involved, and in which order, leading to the universal genetic code. That code is one of a tiny number of near optimal codes that have the dual properties of high redundancy and resistance to mutation [108]. It therefore must have been discovered by a process of feedback-constrained symmetry-breaking phase transitions, or “boot-strapping”. The underlying necessity for these transitions are discussed in detail elsewhere [5,6]. Among the first of these symmetry-breaking transitions relevant to the genetic code was the aaRS class division that divided the amino acids into two distinct classes, as discussed in §IV.
III. Structural Biology of Ancestral Synthetases
Most of what we know about the structures of Urzyme and Protozyme models for ancestral aaRS has been inferred from crystal structures of full-length enzymes. Thus, for example, all structures depicted in figures herein were prepared by excerpting relevant coordinates from the corresponding pdb files. However, work has begun on the challenging task of providing more reliable and detailed structural data.
TrpRS Urzyme is a catalytically active molten globule
Attempts to crystallize Urzymes, either alone or as maltose-binding-protein fusions, has not yet been successful. It has, however, been possible to prepare isotopically labeled samples of active TrpRS Urzyme. Preliminary 15N-1H HSQC spectra from those samples supported an unexpected, but not unprecedented conclusion—the TrpRS Urzyme is not a folded protein, but has many of the properties expected from a catalytically active molten globule [64]. The HSQC spectrum has a reasonable dispersion of values in the 15N dimension, but all ~80 peaks are contained between 8–8.5 ppm in the 1H dimension, which is characteristic of proteins that are not fully folded. The conclusion that it is probably a molten globule is reinforced by the fact that the temperature dependence of CD spectrum exhibits cold denaturation, and that Thermaflour measurements of thermal melting in the presence of Sypro Orange dye exhibit high fluorescence at all temperatures below ~45 C, above which fluorescence decreases non-cooperatively over a range of ~30 degrees C. Both cold denaturation and high fluorescence over broad temperature ranges in the presence of Sypro Orange are characteristic of limited tertiary structure as in molten globules [64].
The likely possibility that the TrpRS Uryzme is a catalytically active molten globule has considerable significance in light of the work of Hilvert [109] and Hu [110], showing that the native chorismate mutase dimer and an engineered monomeric form that is a molten globule exhibit comparable rate accelerations—and hence transition state stabilization—by distinctly different strategies. The high, positive TΔS‡ implies even greater enthalpic contribution to transition state stabilization. Thus, thee molten globular catalyst achieves a substantially higher – ΔH‡ to overcome the entropic cost of restricting the molten conformation of the catalyst when it binds to the transition state. The additional flexibility of the molten globular ensemble appears enable it to wrap more tightly around the transition state configuration of the substrate than can the properly folded native form of the enzyme.
The potential manifold for catalytic activity by poorly structured polypeptides may thus be much larger than was thought possible
Two molten globular polypeptides therefore exhibit high rate accelerations. At least one of these does so by forming substantially tighter bonds in the transition state than are possible with the native enzyme. This plasticity opens the possibility that many similar structural ensembles might act catalytically, and hence that a wider range of polypeptides might exhibit catalytic activity.
Peptide catalysts are far superior to ribozymes
The superiority of peptide catalysts is widely recognized. However, because it is so much easier to generate and select catalytic aptamers from RNA than it is from protein combinatorial libraries, it is unclear that this wide recognition comes with an appreciation of just how superior polypeptide catalysts are, in principle. Wills [49] has compared the combinatorial possibilities for making an active site with proteins to those available for ribozymes. The combinatorial advantage of protein active sites arises because amino acids are only half the volume of nucleotide bases, meaning that contact to a transition state can arise from a greater number of amino acids. This advantage is compounded by the fact that there are five times as many choices of amino acids. As a result, proteins have an advantage somewhere between a million- and a billion-fold over RNA, which is what is observed experimentally [5].
Hecht’s work [111–113] has demonstrated that a large proportion of molecules within patterned combinatorial libraries actually do form molten globules. The relatively low free energy barriers associated with assuming catalytically competent conformations, together with the vastly enhanced abilities of amino acid side chains to engineer nanoscale chemistry argue that catalytic activity is likely to arise and evolve much more rapidly from populations of peptides than from libraries of RNA. Thus, demonstrating that catalytic proficiency does not require the evolution of properly folded proteins represents a considerable expansion in their potential catalytic repertoire.
IV. The Basis for the AARS Class Division
Because they activate amino acids by catalyzing adenylation by ATP, the aminoacyl-tRNA synthetases are arguably among the earliest enzymes to emerge during the origin of life. Absent catalysts, amino acid activation is both the slowest kinetically and most irreversible thermodynamically of the chemical reactions necessary for protein synthesis [34]. The former distinction means that activation is ~1000 times slower than acyl transfer to tRNA or peptide bond formation from activated intermediates and represents the principal kinetic barrier to making peptides in a pre-biotic context. The latter means that amino acid activation is one of the hardest reactions in biology to drive to completion. It is probably not accidental that it became driven by ATP hydrolysis, which can deliver an additional free energy pulse once the pyrophosphate liberated by amino acid activation is subsequently hydrolyzed, assuring that activation goes to completion.
That two distinct protein superfamilies emerged to couple amino acid activation to ATP hydrolysis represented a conundrum that remained unanswered until a quite recent investigation connecting coding properties of tRNA bases with the physical chemistry of amino acid side-chain phase transfer and protein folding equilibria [40,41,55] provided the first clues to a possible answer (see Fig. 2). One puzzling aspect of dividing the 20 canonical amino acids into two distinct groups is that the resulting classes appear to have quite similar diversity in their representation of the various physical chemical properties. Subclass B activates Glu, Gln, and Lys in Class I and Asp, Asn, and Lys in Class II. Subclass C activates Trp and Tyr in Class I and Phe in Class II. The similar diversity within each class leaves open the possibility that the two synthetase Classes appeared sequentially and not simultaneously. Although most authors have been reluctant to comment on their order of appearance [1], several have argued for a sequential appearance [114–116].
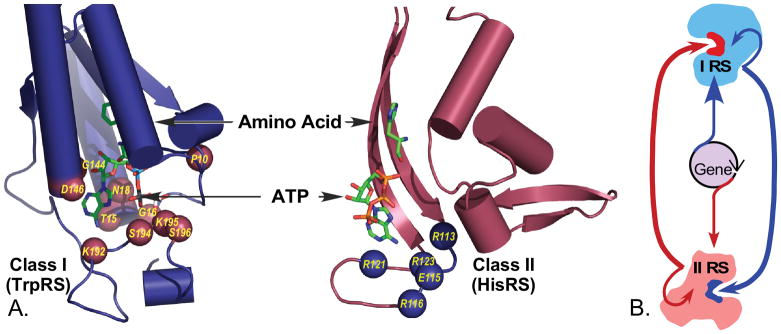
A. Class I, II aaRS have highly interdependent active-site constructions
Sequence conservation within the synthetase active sites furnishes the strongest evidence that the two Classes appeared simultaneously and not sequentially [42,34]. Functional residues in each site—those whose functional groups directly influence the chemistry of the two substrates, as opposed to side chains within the conserved signatures that interact with the rest of the protein—are drawn entirely from the set of amino acids activated by the opposite aaRS Class (Fig. 11). This phenomenon is especially conspicuous for the Class-defining signature residues. For example, seven residues from the HIGH and KMSKS motives of Class I active sites interact with the ATP substrate; whereas the two remaining hydrophobic, Class I, I and M residues, respectively, coordinate movement of the two signatures because they are embedded in a hydrophobic core of the anticodon-binding domain.
Figure 11.
Functional active-site residues in all members of Class I are drawn exclusively from the set of amino acids activated by Class II aaRS, and vice versa. A. The two sets of Class-defining residues have quite distinct functional groups; Class I active-site residues consist of histidine, glycine, proline, asparagine, threonine, lysine, and aspartic acid; Class II active-site residues consist of arginine and glutamic acid. It is highly inconceivable that this differentiation is accidental. B. The differentiation of active-site catalytic residues effectively creates a hypercycle-like interdependence of Class I and II ancestral forms [62].
Although further work on this question is certainly worthwhile, there appear to be functional reasons why active-site residues are deployed quite differently in each Class. Although these differences have not been delineated systematically, they appear to produce dissimilar transition-state stabilization mechanisms [117,118]. With some exceptions—TrpRS residue D132 is a Class II amino acid conserved in the amino acid substrate binding sites of several Class I aaRS—the residues that obey this particular asymmetry are located in the respective ATP binding sites, and their functional differentiation is likely to be related to functional differentiation in the mechanism for ATP activation. Class I aaRS use a dynamic Mg2+ ion that moves with the PPi leaving group bound by the KMSKS loop during the transition state [89] and appear to stabilize additional negative charge in a dissociative transition state involving an α-metaphosphate [89]. Class II aaRS appear to stabilize a pentavalent α-phosphoryl group transition state with multiple Mg2+ ions that are bound directly by protein residues in a manner reminiscent of the two-metal transition state stabilization of polymerases [119].
The only variation in these patterns occurs in eukaryotic synthetases, which are much more highly differentiated than bacterial aaRS and have adapted their catalytic mechanisms to accommodate and perhaps to sustain the accumulation of modules, called physiocrines [120,121], with additional functions, whose selective advantages have been proposed to require modifying the active-site configurations [122,123].
These putative mechanistic differences are consistent with the differential use of Class II histidine, asparagine, lysine and serine—stabilization of a (PO3−) metaphosphate transition state—by Class I aaRS and of Class I arginine—stabilization with Mg2+ of pentavalent phosphoryl transition state assisted by glutamic acid coordination of Mg2+—by Class II aaRS. As the active sites are almost certainly the oldest parts of an enzyme, it seems highly unlikely that either aaRS Class could ever have managed without the other because of the necessity to provide activated amino acids from the opposite set for their own translation.
Mechanistic differentiation as outlined here may have deeper evolutionary significance in light of a series of asymmetries at primary, secondary, and tertiary structural levels. These are described in §IV.B, C, and D.
B. Amino acid side chain volume may underlie the class distinction
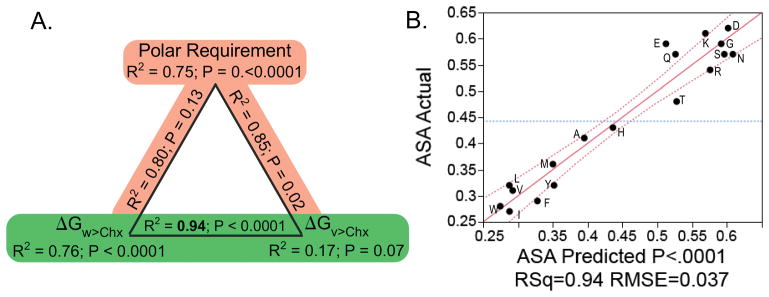
It seems reasonable to seek a basis for the striking division between the two amino acid classes from among the various physical descriptors that differentiate amino acid chemical behaviors. The obvious diversity within each class complicates the question. Given a matrix with one amino acid per row and a column giving its class and additional columns for each candidate descriptor, one can compare the various linear models relating properties to amino acid Class. Most proposed predictors are corrupted by attempts to impose correlations with the buried (or exposed) surface areas in proteins. Three predictors stand apart from this difficulty: the “polar requirement” [124], and the phase transfer free energies for water-to-cyclohexane [125–128] and vapor-to-cyclohexane [129]. The former resulted from an ingenious attempt to test the hypothetical correlation between an amino acid property and the codon table. The resulting scale was derived from paper chromatography of the amino acids in solutions of varying content of dimethylpyridine, to alter the mobility in accordance with the phase transfer behavior. However, that scale is highly idiosyncratic and cannot be recapitulated without an extensive investigation into the properties of paper chromatography, which is rarely if ever used today. In contrast, both the latter measures represent pure physico-chemical equilibria unrelated to possible interactions either with carbohydrates in the paper support or with nucleic acids (Fig. 12).
Figure 12.
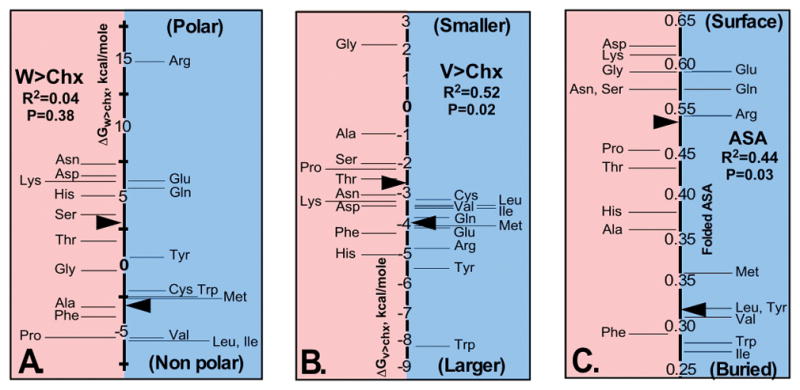
Comparisons of the phase transfer free energies of the Class I (blue) and II (red) amino acids. A. Water to cyclohexane transfer free energies, or hydrophobicity. B. Vapor to cyclohexane transfer free energies, which are closely related to side chain volume (R2 = 0.88). C. Exposed surface areas of amino acid side chains in folded proteins, estimated by Moelbert [130]. Median values are indicated by black arrow points. Statistics for regression models expressing each property as a function of amino acid Class are given below the titles. Extremes for each property are indicated at the top and bottom of the Class I half of each panel. (Adapted from Carter, CW Jr & Wolfenden, R. (2015) Proc. Nat. Acad. Sci. USA 24: 7489–7494.)
The three different properties are not linearly independent. The polar requirement is uncorrelated with the vapor-to-cyclohexane equilibria (R2 = 0.06; P = 0.26) but well-correlated with the water-to-cyclohexane equilibria (R2 = 0.62; P < 0.0001). However, the two phase transfer equilibria themselves are uncorrelated (R2 = 0.01; P = 0.69). Thus, they are distinctly different metrics, whereas the polar requirement resembles the hydrophobicity measured by the water-to-cyclohexane transfer equilibrium.
The second relevant point is how the three values correlate with the degree of side chain exposure in folded proteins measured by the solvent accessible surface area, ASA. That metric is, itself, subject to uncertainty as discussed elsewhere [41,55], where it is argued that the values published by Moelbert [130] represent the least ambiguous values. Moreover, amino acids cysteine and proline violate all characterizations of this sort, owing to alternative influences—exposure in turn segments, coordination of metals and disulfide linkages—that lead to highly variant distributions between surface and core. Given those assumptions, and by a substantial margin, the best model for the ASA values is achieved by a linear combination of water-to-cyclohexane and vapor-to-cyclohexane free energies (R2 = 0.94; all P < 0.001). The polar requirement performs less well in combinations (Fig. 13), indicating that the information in the polar requirement is redundant with that in the transfer free energy from water to cyclohexane. Thus, the vapor-to-cyclohexane transfer free energy provides new, complementary information, allowing a nearly complete prediction of ASA.
Figure 13.
Basis sets for protein folding. The polar requirement [124] was the earliest effort to find such a basis set. Performance of the polar requirement is compared here with two other, more fundamentally derived amino acid properties. A. Summary of regression models for the residual accessible surface area in folded proteins, ASA, [130] using amino acid properties as predictors. All three metrics are related to the ASA. Polar requirement alone is correlated with ASA nearly as well as is the water to cyclohexane transfer free energy, to which it is highly correlated. Edges of the triangle give statistics for bi-variate models involving two of the three predictors. Green background indicates satisfactory models; red background indicates models that have inferior statistics, either because they do not explain the variation in ASA, or because the polar requirement contribution is insignificant, or both. B. The optimal model given across the bottom edge of A leads to quite satisfactory prediction of the ASA for all canonical amino acids excepting proline and cysteine, for which there are consensus factors leading to their being outliers.
The original purpose in developing the polar requirement was to account for regularities in the genetic code. It appears, however, that tRNA identity elements are more reliably related in detail to the phase transfer free energies [40,41] than to the polar requirement value.
C. tRNA acceptor stem and anticodon have independent coding properties
In a paper that now appears increasingly prescient, Schimmel, et al. [23] argued that the dual domain structures of aaRS and tRNAs and experimental demonstrations that many aaRS could specifically aminoacylate tRNA acceptor stems in the absence of the anticodon stem loops implied an earlier phase of genetic coding. They proposed that aaRS catalytic domains and tRNA acceptor stems may have implemented an “operational RNA code” that enabled them to begin to align aminoacyl-acceptor stems according to an ancestral messenger RNA [131]. The anticodon stem-loop and corresponding binding domains in the synthetases were assimilated later.
The more recent demonstration that aaRS Urzymes could acylate tRNA complemented the experimental acylation of acceptor stems [132–134], reinforcing the suggestion of Schimmel, et al., and highlighting the question of what, specifically, might that operational code have consisted? To answer this question, various potential properties of the 20 canonical amino acids were assembled into a table with one line per amino acid. Each property was listed in a separate column. Separate tables included additional columns representing acceptor stem and anticodon bases forming identity elements (compiled by Giegé [135]) according to a binary code using one bit for whether a base is a purine (−1) or a pyrimidine (1) and another bit for whether its Watson-Crick base pairing formed three (1) or two (−1) hydrogen bonds. Regression models were then constructed for each property as a dependent variable using the coding element columns as independent variables [41].
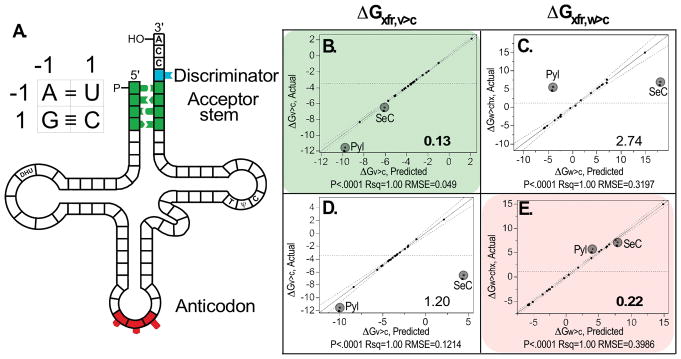
Not surprisingly, all such models provided excellent correlations with each physical property if sufficiently many coefficients were used. To differentiate “predictive” models from models using sufficiently many coefficients that they overfitted the noise, two non-canonical amino acids, selenocysteine (Sec) and pyrrolysine (Pyl), outside the training set were used for cross-validation. There was a clear distinction between models capable of predicting the properties of the two amino acids in the test set, and those that did so poorly. “Predictive” models had a small variance in predicting the test set (Sec, Pyl). They differed distinctly, depending on whether the identity elements used as independent variables came from the acceptor stem or from the anticodon (Fig. 14). Bases in the acceptor stem provide uniquely predictive codes for the side-chain size, whether or not it is branched at the β-carbon, and whether or not the side chain has a carboxyl group. Most other side-chain properties, notably including the hydrophobicity, are specified by the anticodon [41].
Figure 14.
Coding assignments in the tRNA acceptor stem and anticodon bases [40,41]. tRNA coding of amino acid properties. (A) Binary representation of tRNA coding showing the acceptor stem (green) and anticodon (red). (B–E) Correlations between experimental values for ΔGw>c (C and E) and ΔGv>c (B and D) and those calculated from the best regression models. B and C show models for the acceptor stem bases; D and E show models for the anticodon bases. B–E are arranged as a 22 factorial design for the two tRNA coding regions (down the vertical) and the two physicochemical properties (across the horizontal). Coefficients trained on the 20 canonical amino acids ([41]; see SI Appendix, Tables S2 and S4) were used to predict values for Sec and Pyl for cross-validation. Lower right-hand corners of B–E show RMS relative errors for cross-validation. Colored backgrounds show which of the four models make low variance predictions for the test-set of selenocysteine and pyrrolysine, which were not included in the training set. Plots prepared using JMP [182]. (From Carter, CW Jr & Wolfenden, R. (2015) Proc. Nat. Acad. Sci. USA 24: 7489–7494.)
The unique and restricted coding properties of the acceptor stem provide substantive support for the proposal that an “operational RNA code” preceded the universal genetic code carried by the anticodon bases. Details of the acceptor stem code suggest in addition that the properties most important for that code were size, β-branching, and carboxylate side chains. In turn, those properties argue that genetic coding began before it became useful to encode side chains necessary to form hydrophobic cores, and hence before coding specified folded tertiary structures [41]. In fact, the central features of the acceptor stem code specify requirements for forming extended chain β-structures, like those identified by modeling interactions between peptide β-structures and RNA [136,137]. These requirements suggest a selective advantage that could have favored the emergence of such an operational code from a pre-existing population of oligopeptides and oligonucleotides that interacted according to a direct, stereochemical code based upon mutual structural complementarity. Moreover, it is consistent with the notion, elaborated in §II.D and §V.B, that ancestral synthetases, and especially protozymes were coded using a reduced alphabet, leading to “statistical proteins” [138–140,49].
D. Bi-directional coding implies two interpretations of the same genetic information
To test the Rodin-Ohno hypothesis directly, we adapted the Rosetta multistate design algorithm [141] to craft polypeptide sequences to stabilize two alternative backbone configurations—Class I and Class II protozymes— simultaneously and subject to the constraint that amino acids selected to stabilize one backbone have codons complementary to those of amino acids at the corresponding position on the other strand. Those constraints enforced bi-directional coding and produced one gene from which we could express a Class I Protozyme in one orientation and a Class II Protozyme in the other orientation Fig. 15 [26].
Figure 15.
A designed bi-directional gene encoding a Class I Protozyme on one strand and a Class II Protozyme on the opposite strand [26]. A. Structural scaffolds that constrained Rosetta amino acid selections while enforcing complementary codons on opposite strands. Scheme in the center indicates locations of substrate binding sites. B. Translated sequences from the designed gene. Alanine mutations used to validate authenticity of activity are highlighted in red above the corresponding active-site residues. Zoomed region illustrates complementary coding sequences. (This research was originally published in the Journal of Biological Chemistry Martinez, L. et. al., (2015) Journal of Biological Chemistry 290:19710–19725 ©American Society of Biochemistry and Molecular Biology.)
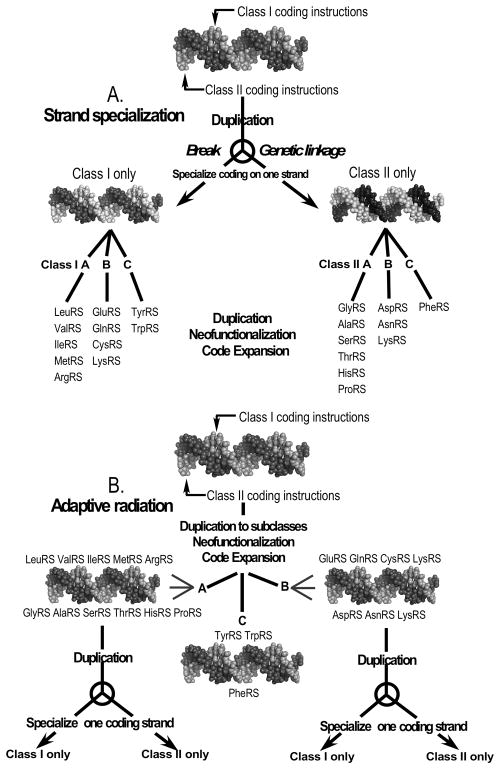
Contemporary aaRS genes are obviously coded uni-directionally (there is, however, evidence that bi-directional coding might have survived to contemporary organisms in isolated cases [142–144,48]. The degree of middle codon-base pairing in sufficiently detailed, all-by-all comparisons may therefore allow two different mechanisms to be distinguished: (i) strand specialization, in which the two strands of daughter genes developed mutations that eliminated bi-directional coding in order to achieve sufficiently improved fitness (Fig. 16A), and (ii) adaptive radiation of bi-directional genes that would have preserved high middle codon-base pairing until more recent times (Fig. 16B). At what point the strand specialization actually occurred along the sequence of bifurcations during code expansion should have left distinguishable signatures in patterns of the middle codon-base pairing distance metric.
Figure 16.
Alternative mechanisms for genetic code expansion. A. Strand specialization. Daughters produced by gene duplication evolve first by breaking the genetic linkage between bi-directional coding sequences, producing specialized genes that express from only one of the two strands. B. Adaptive radiation. The code is expanded by producing pairs of bi-directional genes, both strands of which have specialized functions. (A) and (B) should lead to different patterns in the degree of middle codon-base pairing. (Adapted with permission from the Society for Molecular Biology and Evolution. Chandrasekaran, SN et. al. (2013) Mol. Biol. Evol. 30: 1588–1604.)
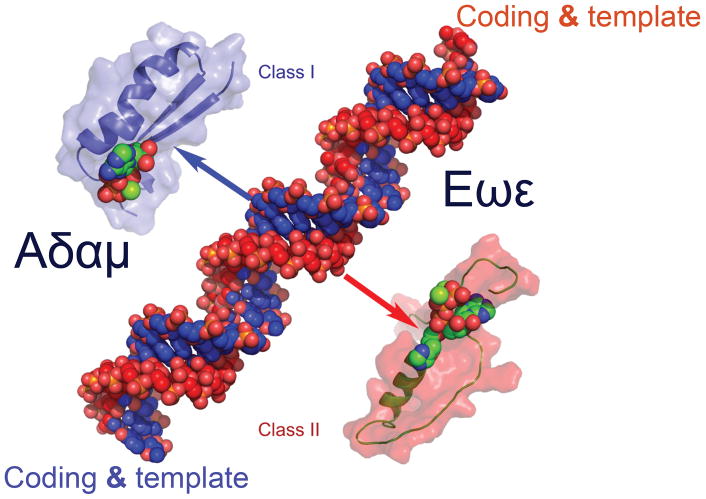
Bi-directional coding means that the two aaRS Classes are derived from alternative interpretations of the same ancestral genetic information, much as in visual puzzles with complementary interpretations of figure and ground (Fig. 17). The Watson-Crick base-pairing rules and the repeating two-fold symmetry relating backbones of opposite nucleic acid strands means that the two strands of the designed Protozyme gene contain only one set of unique information, represented in complementary forms by either strand. The opposite strand has no additional information! Yet that information can support two entirely different interpretations with similar functions, depending on how it is read. This unexpected duality unifies the two aaRS superfamilies. Unification is commonly sought by physicists, but is very unusual in structural biology.
Figure 17.
Two different interpretations of the same unique information. A. The bi-directional Protozyme gene in Fig. 14 has two possible translation products, one from each strand. One double-stranded gene constructed by the computer program “Rosetta” produces two different, equally functional protozymes from instructions on opposite strands. Each strand thus serves as a gene, coding for a peptide, as well as a template for duplication. We have chosen to call the two strands Aδαμ and Eωε, using Greek names to distinguish these molecular ancestors from our mythical human ancestors. Although there is only a single unique set of instructions, that information has two distinct and functional interpretations, depending on which strand is read. (Courtesy of Carter Natural History CW, Jr. (2016) Natural History 125:28–33.) B. This duality is a biological implementation of the familiar visual puzzle, which also has two distinct and equally valid interpretations (Gianni Sarcone, with permission).
V. Inversion symmetries in structure and function maximally differentiate the two aaRS Classes
Anomalies described in §IV appeared at first to be unrelated curiosities. In fact, however, they all assume coherent interpretations as inversion symmetries in the structural and functional implications of bi-directional coding for the organizational levels —primary, secondary, tertiary—of the familiar Linderstrøm-Lang [145] hierarchy. Moreover, these inversion symmetries may have significantly impacted the emergence of genetic coding [5]. This section incorporates these relationships into a unified framework: multi-level molecular disambiguation.
The genes of bi-directionally coded ancestral Class I and Class II aaRS were as different as possible from each other
Although the sugar-phosphate backbones of two complementary strands of a nucleic acid can be interconverted by two-fold symmetry operations, their sequences can be interconverted only via the complementarity operation of base-pairing. This means that the mutational path from one strand to its complement is as long as possible: bi-directionally coded genes are maximally differentiated with respect to mutation, and it is essentially inconceivable that random mutational events could achieve that interconversion.
Primary amino acid sequences of ancestral Class I and Class II aaRS were as different as possible from each other
Primary structures of proteins are intimately related via the genetic code to their nucleic acid (RNA or DNA) coding sequences. For this reason, ancestral, bi-directionally coded aaRS ancestors are maximally differentiated from one another.
Secondary structures of Class I and II aaRS were similar to each other
Unlike primary and tertiary structures, the ancestral Class I and II aaRS secondary structures were very likely similar, with crucial differences outlined in the next paragraph. Secondary structural similarity arises from the fact that formation of α-helical and extended β-structures is driven largely by periodic patterns of similar side chain properties—heptapeptide repeats of non-polar side chains forming α-helices and alternating dipeptide patterns forming extended β-structure. Such periodicities reflect across from one coding strand to the other [See Fig. 6B of ref 7].
Tertiary structures of proteins coded by bi-directional genes have the interesting property of being in a real sense inside out, one to the other
A curious feature in the organization of the genetic code [146] means that amino acids that are confined to cores of folded proteins have codons whose anticodons encode residues invariably found on the surface [See Fig. 6A of ref 7]. Thus, whereas secondary structures in bi-directionally coded pairs of proteins are largely reflected across the two coding strands, solvation patterns of their respective side chains are inverted. Surfaces of helices and strands that are exposed in folded structures of one Class are buried in those of the other.
Use of side chains from opposite Classes maximizes differentation of catalytic mechanisms
The active site differentiation (Fig. 11) results in significant mechanistic disambiguation of the active-site chemistries of Class I and II aaRS. Mutations that might lead to interconversion from one mechanism to the other would therefore be most likely to be lethal, adding a measure of secuity to the mechanistic integrity of the Class.
Substrate recognition differentiates large from small amino acid side chains
The only significant aspect of the canonical 20 amino acids for which there is a statistically significant difference between the two classes is the size of the side chain [41,55]. It would seem more than coincidental that amino acid size is also the primary source of differentiation in the tRNA acceptor stem operational code [40,41]. Side chain volume thus appears to underlie the initial differentiation between aaRS substrates that eventually enabled the development of the current genetic code.
VI. Evolutionary Implications
The genetic code represents one of the deepest unsolved puzzles associated with the origin of life. An important possible reason why it has remained such an outstanding problem is that much of the interdisciplinary community concerned with the origin of life has been preoccupied by the RNA World hypothesis, under which the code remains an even more challenging enigma than it needs to be. Under that hypothesis, the code was “discovered” by ribozymes seeking to improve their severely limited catalytic potential and was perhaps assisted by stereochemical complementarity of nucleotide triplets whose structures were complementary to those of amino acids, for which they eventually became (either) codons or anticodons [147,148]. The typical argument [149] is that amino acids were first recruited by ribozymes as “co-factors” that enhanced the diversity and proficiency of ribozymes. As their superior catalytic functions became manifest, increasingly polymerized forms of amino acids arose through some form of selection, leading to the translation system now used throughout biology. One of many substantial problems with this narrative is that there really is no rational mechanism for bootstrapping the code into existence in an RNA world [5].
This review promised a dialectical survey of ways in which recent advances in understanding the evolution of the aminoacyl-tRNA synthetases have tended to refute the contention that these enzymes “…did not shape the codon assignments”. A substantial number of unexpected results have been put in evidence since that judgment was formulated. Do those observations contribute to “a credible scenario for the evolution of the coding principle itself and the translation system”? Remarkably, the answer appears to be substantively affirmative as outlined in greater detail elsewhere [5,6]. In brief, each of the unexpected oddities arising from recent aaRS research now can be seen as necessary and sufficient vestiges of a bootstrapping process that began deep in a very primitive RNA/peptide world that prefigured relationships in Fig. 2 at an elemental level. Its built-in and robust reflexivity enabled a massive acceleration of the search for a near optimal genetic code. Further, these results were accomplished using a variety of effective new tools and ways of thinking, with which to explore the many interstices remaining to be explored.
Assembling the pieces reviewed here into a coherent refutation of the RNA World narrative begins with the conclusion that contemporary protein structures represent an archive of their origins and evolutionary expansion.
Protein structures were probably not completely overwritten but left an interpretable archive of their evolutionary origin in bi-directional genetic coding
Successive experimental deconstruction of both Class I and Class II aaRS reveals parallel structural and catalytic hierarchies. Catalytic domains, Urzymes, and Protozymes represent increasingly deeply, broadly conserved motifs. The relative ease in identifying them and the robustness of their experimental characterization compose a substantial string of evidence that the hierarchies apparent in Figs. 3 and 4 represent not a palimpsest of an RNA World [150], but a legitimate archive [151]—evident in the contemporary structures—from which we have been able to read out, and reconstruct reasonable models for successive stages in their evolution.
Evolutionary reconstruction is kin to similar scientific problems, such as chemical and enzyme kinetic mechanisms, that present formidable barriers having to do with limitations placed by time. Kinetic mechanisms imply limiting structures, transition states, which by virtue of their extremely short lifetimes, cannot normally be directly detected. The inability to re-play the tape of evolution, on the other hand, creates analogous problems. In both cases, direct study of the objects of greatest interest is either difficult or impossible. These time-related barriers, in turn, dictate the logical framework necessary to progress. Koch’s postulates concerning infectious disease were an early expression of this framework, and these were re-formulated by Fersht [152] to shape the evidence for a particular intermediate in a reaction path. This logical process entails three elements: characterization of the putative intermediate, demonstration that it can be formed from preceding structures fast enough to lie on the path, and demonstration that it can react fast enough to lie on the path. By analogy, a legitimate evolutionary intermediate must be identified, and prepared. Then, plausible evolutionary changes must be outlined—and if possible tested—to account for its initial appearance and its subsequent conversion to the next intermediate in a reasonable timeframe.
The aaRS superfamilies behave consistently with this logical framework. Like Matryoshka dolls, elemental Class I and II active sites both lie within their protozymes, which form the ATP binding sites. Protozymes themselves compose a bit less than half the Urzymes, which contain the nuts and bolts of the catalytic domain. The function of the TrpRS Urzyme within the full-length enzyme is, however, entangled with coordinated motions of the CP1 and ABD domains [153,154,94,155,156]. Recent combinatorial perturbation studies have implicated these motions in long-range allosteric effects crucial to both catalysis [93,157] and specificity [89,32,33] in TrpRS. The synthetase phylogenies therefore represent rich and, as yet only minimally tapped, resources of information about the evolutionary development of catalysis, specificity, and allostery.
Beginning with the audacious proposal of Rodin and Ohno [43] and culminating with the experimental demonstration of a bi-directional gene for the Class I and II Protozymes [26], the trajectory of aaRS research has produced diverse experimental and bioinformatic confirmations of the predictions of bi-directional coding (Fig. 1). Substantive validation of the unification of the two synthetase Classes implies a need to re-annotate proteomic databases. In particular, if, as appears to be the case, a bi-directional synthetase protozyme gene preceded most of the proteome, then phylogenies [158–162] that imply that the Class I synthetase superfamily branched off a different root quite late in the emergence of the proteome cannot also be correct unless modified in important ways to reflect the substantially finer modularity of proteins in general and the staged appearance of different modules.
For the moment, suffice it to say that the direct evidence for the bi-directional coding hypothesis appears exceptionally strong both for Protozymes [26] and for Urzymes [28]. §III developed the implications of bi-directional coding for the differentiation of the genes themselves and their implications for primary, secondary, and tertiary structures of the corresponding synthetases, and thence for their catalytic and coding functions. In the following section, we see how these aspects of synthetase phylogeny and structural biology, viewed coherently as outlined in §V, also in fact fulfill important requirements for bootstrapping complexity from simplicity.
B. Bi-directional coding was probably essential to stabilize the emergence of translation
From the standpoint of information processing, genetic coding differs fundamentally from replication. It is, arguably, one of the most significant and puzzling among the transformations that produced biology from chemistry. It is perhaps the key event that enabled the creation of a multiplicity of sufficiently tunable catalytic activities to synchronize the rates necessary for cellular metabolism, §II.A. It seems surprising that Woese, et al. [2] would have summarily dismissed the possibility that the evolution of the aminoacyl-tRNA synthetases—the executors of the genetic code—had much to do with the development of the code itself.
The alternative argument, that synthetase evolution actually was the sine qua non of what generated the code, has been articulated in considerable detail elsewhere [5,6, and refs cited therein]. Key aspects of that argument are as follows:
Primary structures generated by bi-directional coding are maximally different from one another, hence cannot be “fused” via functional mutants.
Coding relationships in tRNA acceptor stems and anticodons are consistent with at least two distinct stages during which synthetase-tRNA recognition participated in indirect (i.e., “genetic”) coding [40,41]. The earlier stage is eminently consistent with a very small amino acid alphabet consisting of one or at most two bits. Moreover, this result implies the tetrahedral network (Fig. 2) connecting four nodes, two in the nucleic acid world—tRNA (the programming language) and mRNA (the programs) to two nodes in the protein world—amino acid physical chemistry and protein folding [40,41,55].
A two-bit alphabet competing with anything pre-existing in any RNA coding world having higher sophistication would have been rapidly eliminated by purifying selection because it would degrade more sophisticated messages.
(a–c) imply that genetic coding must have been built from scratch in an implementation executed by protein aaRSs.
Bi-directionally coded aaRS Protozymes and Urzymes represent a credible origin of the reflexivity necessary for efficiently bootstrapping the full genetic code into existence.
This bulleted list correlates with the elements of inversion symmetry relating the two aaRS Classes suggesting that the fundamentals of aaRS evolution (§III) closely match requirements for the emergence of genetic coding (§V.B(a)–(e)).
Bi-directional coding is unexpected because it limits genetic diversity
An indeterminate, but presumably significant fraction of all possible mutations that might enhance the fitness of both products from a bi-directional gene are forbidden by the complementarity constraint. It also appears likely that many such mutations would also decrease the fitness of the translated product from the opposite strand. Recent unpublished computational analysis [163] suggests that the cost of bi-directional coding may be smaller than previously thought. Computational construction of bi-directional genes for all pairwise combinations of 500 pfam domains revealed that the number of pairs whose bi-directional genes in all 6 relative reading frames were homologous to consensus sequences from contemporary sequence alignments was unexpectedly high. Thus, the inversion symmetry of the universal genetic code observed first by Zull and Smith [146] may actually have played a key role in the eventual selection of the universal genetic code by optimizing the number of potentially functional bi-directional genes.
In any case, co-linear bi-directional coding does exact a price. The quest for diversity is severely limited by constraining both strands of a gene to have functional interpretatinos. That price must have been paid for by strong contemporary selective advantages. Two definitive properties—gene linkage [68] and gene differentiation [5,6]—together with the interdependence of all aaRS genes, may have compensated for this limitation while error rates were very high, and consequently made bi-directional coding an inevitable requirement for the emergence of coded protein synthesis.
Bi-directional coding links gene expression
It seems reasonable that the two classes of amino acid substrates differed sufficiently that coded protein synthesis could not have been launched without (at least) two specialized kinds of aaRS. The distinction between amino acid substrates of Class I and II aaRS is closely related to the roles those amino acids play in protein folding and the apparently earlier distinctions based on size, β-branching, and carboxylate side chains [40,41] are conspicuously appropriate for defining secondary structures in coded peptides. Bi-directional coding of amino acid activating enzymes with two specificities would have created a durable and ready-made basis for beginning to define coded peptides with rudimentary functional distinctions. If so, then before amino acid activation and acylation reactions became compartmentalized, bi-directional coding would have linked the two gene products, ensuring that both kinds of aaRS were produced in the same places and at the same times.
Bi-directional coding assures the stability necessary to initiate and sustain genetic diversity
More generally, the inversion symmetry of bi-directional aaRS gene coding fulfills a number of criteria necessary for the stability and survival of emerging quasispecies in the face of what have come to be called “error catastrophes” [164,62,165,149]. The relatively low fidelity of the Urzymes characterized thus far and the evidence that they themselves are highly evolved mean that their origins lie in populations of molecules that achieve similar functions, but with multiple sequences called “quasispecies” [166]. The centroids of quasispecies are powerful “attractors” because they are sufficiently isolated in sequence space that variants with lower function will eventually be eliminated unless they “revert” toward the centroid. It is difficult therefore for a quasispecies to “bifurcate”. The most important barrier to generating multiple aaRS was therefore to establish two species with similar catalytic function that were sufficiently differentiated that they could form stable, independent quasispecies [See Figure 2 of [5]. Fig. 18 summarizes how bi-directioinal coding provides the requisite differentiation to establish a “boot block” for the self-organization of genetic coding [5,6] in a peptide•RNA collaboration.
Figure 18.
Summary of how the surprising aspects of aaRS evolutionary history actually helped to facilitate the stabilization of quasi-species bifurcations associated with the earliest genetic coding. At each level of the Linderstrøm-Lang hierarchy [145], from coding strands to secondary, to tertiary structure and mechanism. Bi-directional coding decisively differentiates between the two aaRS Classes.
The genetic code is much too unusual to have been discovered by chance
Among the challenges associated with genetic coding is to understand how so much amino acid physical chemistry became embedded into both tRNA and mRNA sequences. The combinatorics of genetic coding have been analyzed multiple ways by multiple investigators. The conclusion of these studies has been that for every code with the properties of the universal genetic code, there are perhaps a million other potential codes that are less optimal [108]. That estimate should probably be revised as both the apparent temporal appearance of encoded amino acid physical chemistry and bi-directional coding impose even more stringent requirements, making the universal code even more special than Freeland & Hurst appreciated.
In any RNA world, selecting amino acid sequences that fold into functional proteins depends entirely on natural selection. This implies an essentially trial-and-error search. As Koonin has pointedly noted by invoking multiple universes, such a specialized code is inaccessible by random processes in our universe [149]. The alternative, which thus appears mandatory, is to provide a bootstrapping algorithm, by which a simple process can be endowed with the necessary characteristics to build complexity using its own resources.
The bootstrapping metaphor is quite intimately embedded into the framework of genetic coding. As noted [40,41], the code comprises a programming language and an associated set of programs written using that language. In this sense, it closely resembles a computer operating system, as Williams has noted elsewhere [52,167,168]. The key here is that computer operating systems are built around a simple set of instructions sufficient to enable the hardware, by executing those instructions, to build successively more sophisticated levels of functionality using a very limited set of alternatives. We believe that genetic coding must have arisen from an analogous “boot block” [5], some of whose key relationships are illustrated in Fig. 19.
Figure 19.
Reflexivity in the emergence of the genetic code. Bi-directional Protozyme and putative Urzyme genes compose an existence proof for an intrinsic coding feedback loop that cannot exist in an RNA world. These genes are potentially translated (thick grey arrows; rules) by themselves (rule executors; thin, dashed arrows). Both genes, in turn, must fold according to the behavior of amino acid phase transfer equilibria (local environment sensing) in order to execute the coding rules (thick black arrows). This feedback loop greatly enhances the ability of such genes to serve as “boot block” in bootstrapping development of more sophisticated genetic coding.
As shown in Fig. 19, bi-directionally coded protein aaRS are uniquely equipped to implement the crucial feedback loop necessary to bootstrap genetic coding, namely sensing the impacts of the local nano-environment on component amino acids that lead to protein folding rules. This feedback loop assures that amino acid sequences incapable of folding or whose functions are inferior are rapidly eliminated because the “rule executors” are themselves governed by the same phase transfer equilibria as all proteins. It cannot operate in any system using ribozymal aaRS. Bi-directional synthetase genes and the coding rules (Fig. 19) therefore together compose an existence proof that genetic coding could have evolved from humble origins by discovering both foldable sequences and optimal coding relationships much more rapidly than would have been possible in a pre-existing RNA world.
Interdependence helped assure survival of both synthetase classes
The bullet list at the beginning of this section differentiates the products of a bi-directional gene at all levels. This multi-level differentiation sustains their underlying functional separation. The probability that mutations could eventually fuse their functions is minimized by the decisive mechanistic disambiguation [see Fig. 2 of Ref 5]. Further, dependence of the functions coded by each strand on the gene products of both strands defines a hypercycle-like coupling [62] to ensure that the two gene products have enhanced ability to survive high error rates, as in Figs. 17,18. In this sense, the liability we see today in bi-directional coding—tight genetic linkage—was probably a significant strength before chemistry became localized in cells.
Catalysis arose from simple, promiscuous molten globules
The progression of transition-state stabilization free energies illustrated in Fig. 4 already suggests that catalytic proficiency developed progressively during the evolutionary maturation of the aaRS. protozymes, with ~50 amino acid residues produce >40% and Urzymes with ~130 amino acid residues produce ~60% of the transition state stabilization of modern enzymes. The specificity spectra in Fig. 5 suggest that aaRS Urzymes had achieved only 20% of their contemporary specificity. That distinction between catalysis and specificity sharply delineates discrete events in their evolutionary history (Fig. 6). Thus, most of the specificity appears to have evolved after the synthetases had developed most of their catalytic proficiency. Preliminary published experiments suggest that amino acid specificity can be achieved only by invoking allosteric interactions between domains in the contemporary enzymes [89,32,33] (vide infra; §II.C).
The surprising catalytic proficiency of the aaRS protozymes suggests that the earliest transition-state stabilization mechanisms arose by positioning backbone binding determinants and only later made use of active-site side chains. A pertinent example is the N-terminal array of four unsatisfied hydrogen bond donors in alpha helices, which became a foundation for stabilizing phosphate—and pyrophosphate—binding [169] and which is an important part of the Class I protozyme. It is not as yet known whether or not specific active-site side chains in the protozymes function in the same manner as they appear to do in the Urzymes and full-length enzymes. The loss of activity in the active-site mutant protozymes suggests that these side chains do contribute to transition-state stabilization, but more detailed functional studies will be necessary to delineate precisely their functional role.
Similarly, it appears likely that the Urzyme level of evolutionary development may utilize tight transition-state binding associated with a large unfavorable negative entropy change as suggested for the TrpRS Urzyme [64] and a chorismate mutase molten globular variant [110]. This possibility, combined with the discussion in §III of the superiority of peptide catalysts suggests strongly that the emergence and selection of catalytic activity itself was vastly more efficient and hence more rapid for peptide, than for ribozymal catalysts. Moreover, if tight transition-state binding is common among molten peptide molten globules, selection would recruit catalysts from a much larger manifold of sequences. The large negative TΔS term may ultimately limit the potential transition-state stabilization free energy of molten globules, and hence create in addition a selective advantage for evolving sequences that stabilize folded structures with more rigid transition-state complementarity [170]. Finally, as noted above, enhancing substrate specificity likely required the emergence of allosteric effects that cannot develop within the Urzyme framework alone.
VII. Remaining questions
Finally, it remains to outline areas that remain unresolved, where future experimental efforts can be most productive. We pose some of these questions in this section.
What was the scale of modularity in the earliest evolution of proteins?
The most serious challenge to the various scenarios described in this review is the significant evidence from Koonin’s group [149,171,159–161] and others [53,172] that speciation of the Class I aaRS did not occur until relatively late in the generation of the proteome. Those studies are based on the most current thinking on phylogenetic reconstruction, yet they appear to be inconsistent with a fundamental role for objects like the bi-directional Protozyme gene products near the root of the proteome. Our view is that this conundrum arises from failure to appropriately recognize fine-scale modularity in constructing trees for domains. Thus, we believe that the putative bi-directional Protozyme gene was ancestral not only to the aaRSs but to many other families of related function, and that horizontal modular transfers may have been important before molecular biology created cellular species [173,161]. Under this alternative hypothesis, the late branching of Class I aaRS represents a subsequent process associated with refinement of the aaRS specificities, once the proteome as a whole had emerged from a simpler alphabet.
Although the evidence implies that we can infer the modular outlines of evolutionary succession from the structural hierarchies, the assembly of contemporary enzymes from these reconstructed ancestral modules necessarily involved overwriting some details that therefore remain speculative. Our view is that the clearest guides to the actual ancestry remains experimental characterization of the functionality of modules clearly related by phylogenetic methods to contemporary proteins, and that the ability now to investigate the experimental recapitulation of intermodular interactions, both in cis and in trans, along lines developed with the TrpRS [33] and HisRS [29] Urzymes remains a key direction for further research, including both the assembly of Urzymes from protozymes and the other modules present in the Urzymes [31] and the assembly of modern aaRS from Urzymes in the presence of homologs of CP1, the Class II insertion domains, and the anticodon-binding domains of both classes.
Recent work appears to be moving toward a consensus consistent with our view. In particular, Caetano-Anolles [174] now recognizes that the P-loop hydrolase domain genes are among the more ancient genes, without acknowledging that the Class I protozyme appears to be a reasonable ancestral form, not only of the Class I synthetases, but also of the P-loop hydrolases themselves.
Was specific aminoacylation of tRNA originally catalyzed by ribozymes?
The aaRS protozymes appear to be efficient catalysts of amino acid activation. It remains, among other things to be tested, to characterize their amino acid preferences. More important, however, is to determine whether or not they can also accelerate the transfer of activated amino acids to tRNA. The Class I protozyme lacks even the rudiments of a surface with which to bind the tRNA acceptor stem, whereas the Class II protozyme does retain such rudiments. Thus, it is possible that the ancestral protozymes were not entirely symmetrical in their catalytic repertoire, and that the Class II protozyme may have been uniquely able to acylate the tRNA acceptor stem. In any case, it appears that it is much more straight-forward to use Selex procedures to isolate RNA aptamers that use activated amino acids to acylate tRNA than it is to find comparable aptamers capable of activating amino acids by reaction with ATP [175,176]. Thus, it is both conceivable and worth testing whether or not if accompanied by protozymes, such aptamers might have accelerated tRNA acylation from amino acids and ATP.
How simple an amino acid alphabet can still support an active bi-directional protozyme gene?
Answers to this question appear to be fundamental to understanding the origins of genetic coding. Fortunately, the wherewithal to answer it appears to be in place. The middle codon-base pairing metric of bi-directional coding appears to show sufficient variation between pairs of Class I and II aaRS Ur-genes (Figs. 9,10) to support a semi-quantitative map of bifurcations that best account for the order in which the amino acids were assimilated into the growing genetic code [7], (Chandrasekaran, personal communication). The BEAST computer program has been adapted to utilize transition probability matrices of decreasing order, consistent with identifying nodes in the elaboration of the code [106], (Wills, personal communication). The multistate algorithm by which Rosetta imposes gene complementarity can be modified for testing models of code development by generating bi-directional protozyme genes whose catalytic activities can then be compared. These tools will become even more useful if and when it becomes possible to design a bona fide Ur-gene having all three of the modules identified originally by Pham, et al. [31].
How did cognate tRNAs evolve to distinguish between the two aaRS Classes?
This question appears to pose a much more difficult problem. The aaRS class distinction was straightforward to identify from the initial observation of a group of aaRSs that lacked the HIGH and KMSKS catalytic signatures characteristic of all aaRS sequences available at that time [177,44]. Notwithstanding the evidence accumulated to date from a much larger tRNA research community, it is not yet possible to identify sequence signatures—outside the identity elements that define the amino acid specificities of the synthetases—that differentiate the recognition by one or the other synthetase Class. Thus, whereas it is possible in principle, even in the face of horizontal gene transfer [178], to attempt to establish a tree along which the aaRS genes radiated to enlarge the amino acid alphabet, no such exercise appears possible yet for their cognate tRNAs. It is possible that the approaches used by Caetano-Anollès [174] are capable of identifying appropriate patterns in the tRNA multiple sequence alignments, but thus far, that does not appear to have been a goal. Thus, a full understanding of the “tree” of amino acid acylation to tRNA in the emergence of translation, even in principle, appears still to lie ahead.
Acknowledgments
Research from the Carter laboratory was supported by the National Institutes of General Medical Sciences, GM 78228 and GM40906. I am grateful for the comments of an anonymous referee, and for similar input from E. First.
Abbreviations
- aaRS
aminoacyl-tRNA synthetase(s)
- TrpRS
tryptophanyl-tRNA synthetase
- LeuRS
Leucyl-tRNA synthetase
- HisRS
histidyl-tRNA synthetase
- ATP
adenosine 5′ triphosphate
- PPi
inorganic pyrophosphate
- ASA
Solvent-accessible surface area
- HSQC
Heteronuclear Single-Quantum Correlation
- BEAST
Bayesian Evolutionary Analysis Sampling Trees
Footnotes
Conflict of Interest
The author is unaware of any conflicts of interest.
References
- 1.Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA Synthetases, the Genetic Code, and the Evolutionary Process. Microbiol Mol Biol Rev. 2000;64(1):202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koonin EV, Novozhilov AS. Origin and Evolution of the Genetic Code: The Universal Enigma. IUBMB Life. 2009 Feb;61(2):99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert W. The RNA World. Nature. 1986;319:618. [Google Scholar]
- 4.Robertson MP, Joyce GF. The Origins of the RNAWorld. Cold Spring Harb Perspect Biol. 2012;4:a003608. doi: 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter CW, Jr, Wills PR. Interdependence, Reflexivity, Fidelity, and Impedance Matching: the Need for an Alternative to the RNA World. BioRxiv. 2017 doi: 10.1101/139139. [DOI] [Google Scholar]
- 6.Wills PR, Carter CW., Jr Insuperable problems of an initial genetic code emerging from an RNA World. BioRxiv. 2017 doi: 10.1101/140657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrasekaran SN, Yardimci G, Erdogan O, Roach JM, Carter CW., Jr Statistical Evaluation of the Rodin-Ohno Hypothesis: Sense/Antisense Coding of Ancestral Class I and II Aminoacyl-tRNA Synthetases. Molecular Biology and Evolution. 2013;30(7):1588–1604. doi: 10.1093/molbev/mst070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cammer S, Carter CW., Jr Six Rossmannoid Folds, Including the Class I Aminoacyl-tRNA Synthetases, Share a Partial Core with the Anticodon-Binding Domain of a Class II Aminoacyl-tRNA Synthetase. Bioinformatics. 2010;26(6):709–714. doi: 10.1093/bioinformatics/btq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buehner M, Ford GC, Moras D, Olsen KW, Rossmann MG. D-Glyceraldehyde 3-Phosphate Dehydrogenase: Three Dimensional Structure and Evolutionary Significance. Proc Nat Acad Sci USA. 1973;70:3052–3054. doi: 10.1073/pnas.70.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classed based on mutually exclusive sets of sequence motifs. Nature. 1990;347(9):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 11.Delarue M. An asymmetric underlying rule in the assignment of codons: Possible clue to a quick early evolution of the genetic code via successive binary choices. RNA. 2007;13:1–9. doi: 10.1261/rna.257607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delarue M, Moras D. Aminoacyl-tRNA synthetases: Partition into two classes. In: Eckstein F, Lilley DMJ, editors. Nucleic Acids and Molecular Biology. Vol. 6. Springer-Verlag; Berlin, Heidelberg: 1992. pp. 203–224. [Google Scholar]
- 13.Ibba M, Francklyn C, Cusack S. MBIU. Landesbioscience; Georgetown, TX: 2005. Aminoacyl-tRNA Synthetases. [Google Scholar]
- 14.Cusack S. Eleven down and nine to go. Nat Str Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 15.Härtlein M, Cusack S. Structure, Function and Evolution of Seryl-tRNA Synthetases: Implications for the Evolution of Aminoacyl-tRNA Synthetases and the Genetic Code. J Mol Evol. 1995;40:519–530. doi: 10.1007/BF00166620. [DOI] [PubMed] [Google Scholar]
- 16.Cusack S. Evolutionary Implications. Nat Struct Mol Biol. 1994;1:760. [Google Scholar]
- 17.Cusack S. Sequence, structure and evolutionary relationships between class 2 aminoacyl-tRNA synthetases: An update. Biochimie. 1993;75:1077–1081. doi: 10.1016/0300-9084(93)90006-e. [DOI] [PubMed] [Google Scholar]
- 18.Ribas de Pouplana L, Schimmel P. Two Classes of tRNA Synthetases Suggested by Sterically Compatible Dockings on tRNA Acceptor Stem. Cell. 2001;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- 19.Ribas de Pouplana L, Schimmel P. Operational RNA Code for Amino Acids in Relation to Genetic Code in Evolution. J Biol Chem. 2001;276:6881–6884. doi: 10.1074/jbc.R000032200. [DOI] [PubMed] [Google Scholar]
- 20.Ribas de Pouplana L, Schimmel P. Aminoacyl-tRNA synthetases: potential markers of genetic code development. TIBS. 2001;26(10):591–596. doi: 10.1016/s0968-0004(01)01932-6. [DOI] [PubMed] [Google Scholar]
- 21.Schimmel P, Ribas de Pouplana L. Footprints of aminoacyl-tRNA synthetases are everywhere. TIBS. 2000;25(5):207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 22.Schimmel P. Origin of genetic code: A needle in the haystack of tRNA sequences. Proceedings of the National Academy of Sciences, USA. 1996;93:4521–4522. doi: 10.1073/pnas.93.10.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schimmel P, Giegé R, Moras D, Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Nat Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schimmel P. Classes of aminoacyl-tRNA synthetases and the establishment of the genetic code. Trends in Biological Sciences. 1991;16(1):1–3. doi: 10.1016/0968-0004(91)90002-d. [DOI] [PubMed] [Google Scholar]
- 25.O’Donoghue P, Luthey-Schulten Z. On the Evolution of Structure in Aminoacyl-tRNA Synthetases. Microbiol Mol Biol Rev. 2003;67(4):550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez L, Jimenez-Rodriguez M, Gonzalez-Rivera K, Williams T, Li L, Weinreb V, Niranj Chandrasekaran S, Collier M, Ambroggio X, Kuhlman B, Erdogan O, Carter CWJ. Functional Class I and II Amino Acid Activating Enzymes Can Be Coded by Opposite Strands of the Same Gene. J Biol Chem. 2015;290(32):19710–19725. doi: 10.1074/jbc.M115.642876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter CW., Jr Urzymology: Experimental Access to a Key Transition in the Appearance of Enzymes. J Biol Chem. 2014;289(44):30213–30220. doi: 10.1047/jbcR114.576495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Francklyn C, Carter CW., Jr Aminoacylating Urzymes Challenge the RNA World Hypothesis. J Biol Chem. 2013;288:26856–26863. doi: 10.1074/jbc.M113.496125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Weinreb V, Francklyn C, Carter CW., Jr Histidyl-tRNA Synthetase Urzymes: Class I and II Aminoacyl-tRNA Synthetase Urzymes have Comparable Catalytic Activities for Cognate Amino Acid Activation. J Biol Chem. 2011;286:10387–10395. doi: 10.1074/jbc.M110.198929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pham Y, Kuhlman B, Butterfoss GL, Hu H, Weinreb V, Carter CW., Jr Tryptophanyl-tRNA synthetase Urzyme: a model to recapitulate molecular evolution and investigate intramolecular complementation. J Biol Chem. 2010;285:38590–38601. doi: 10.1074/jbc.M110.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pham Y, Li L, Kim A, Erdogan O, Weinreb V, Butterfoss G, Kuhlman B, Carter CW., Jr A Minimal TrpRS Catalytic Domain Supports Sense/Antisense Ancestry of Class I and II Aminoacyl-tRNA Synthetases. Mol Cell. 2007;25:851–862. doi: 10.1016/j.molcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Weinreb V, Li L, Chandrasekaran SN, Koehl P, Delarue M, Carter CW., Jr Enhanced Amino Acid Selection in Fully-Evolved Tryptophanyl-tRNA Synthetase, Relative to its Urzyme, Requires Domain Movement Sensed by the D1 Switch, a Remote, Dynamic Packing Motif. J Biol Chem. 2014;289:4367–4376. doi: 10.1074/jbc.M113.538660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Carter CW., Jr Full Implementation of the Genetic Code by Tryptophanyl-tRNA Synthetase Requires Intermodular Coupling. J Biol Chem. 2013 Nov;288(29):34736–34745. doi: 10.1074/jbc.M113.510958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter CW, Jr, Li L, Weinreb V, Collier M, Gonzales-Rivera K, Jimenez-Rodriguez M, Erdogan O, Chandrasekharan SN. The Rodin-Ohno Hypothesis That Two Enzyme Superfamilies Descended from One Ancestral Gene: An Unlikely Scenario for the Origins of Translation That Will Not Be Dismissed. Biology Direct. 2014;9:11. doi: 10.1186/1745-6150-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodin AS, Szathmáry E, Rodin SN. On origin of genetic code and tRNA before translation. Biology Direct. 2011;6:14. doi: 10.1186/1745-6150-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodin A, Rodin SN, Carter CW., Jr On Primordial Sense-Antisense Coding. Journal of Molecular Evolution. 2009;69:555–567. doi: 10.1007/s00239-009-9288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodin SN, Rodin AS. On the origin of the genetic code: Signatures of its primordial complementarity in tRNAs and aminoacyl-tRNA synthetases. Heredity. 2008;100:341–355. doi: 10.1038/sj.hdy.6801086. [DOI] [PubMed] [Google Scholar]
- 38.Rodin SN, Rodin A. Partitioning of Aminoacyl-tRNA Synthetases in Two Classes Could Have Been Encoded in a Strand-Symmetric RNA World. DNA and Cell Biology. 2006;25:617–626. doi: 10.1089/dna.2006.25.617. [DOI] [PubMed] [Google Scholar]
- 39.Rodin SN, Rodin A. Origin of the Genetic Code: First Aminoacyl-tRNA Synthetases Could Replace Isofunctional Ribozymes When Only the Second Base of Codons Was Established. DNA and Cell Biology. 2006;25:365–375. doi: 10.1089/dna.2006.25.365. [DOI] [PubMed] [Google Scholar]
- 40.Carter CW, Jr, Wolfenden R. Acceptor-stem and anticodon bases embed amino acid chemistry into tRNA. RNA Biology. 2016;13(2):145–151. doi: 10.1080/15476286.2015.1112488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter CW, Jr, Wolfenden R. tRNA Acceptor-Stem and Anticodon Bases Form Independent Codes Related to Protein Folding. Proc Nat Acad Sci USA. 2015;112(24):7489–7494. doi: 10.1073/pnas.1507569112. doi: http://www.pnas.org/cgi/doi/10.1073/pnas.1507569112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter CW., Jr What RNA World? Why a Peptide/RNA Partnership Merits Renewed Experimental Attention. Life. 2015;5:294–320. doi: 10.3390/life5010294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodin SN, Ohno S. Two Types of Aminoacyl-tRNA Synthetases Could be Originally Encoded by Complementary Strands of the Same Nucleic Acid. Orig Life Evol Biosph. 1995;25:565–589. doi: 10.1007/BF01582025. [DOI] [PubMed] [Google Scholar]
- 44.Eriani G, Dirheimer G, Gangloff J. Aspartyl-tRNA Synthetase from Escherichia coli: Cloning and Characterisation of the Gene, Homologies of its Translated Amino Acid Sequence with Asparaginyl- and Lysyl-tRNA Synthetases. Nucleic Acids Research. 1990;18:7109–7117. doi: 10.1093/nar/18.23.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cusack S, Härtlein M, Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Research. 1991;19(13):3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cusack S, Berthet-Colominas C, Härtlein M, Nassar N, Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature. 1990;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 47.Duax WL, Huether R, Pletnev V, Langs D, Addlagatta A, Connare S, Habegger L, Gill J. Rational Genomes:Antisense Open Reading Frames and Codon Bias In Short Chain Oxido Reductase Enzymes and the Evolution of the Genetic Code. PROTEINS: Struct Funct Bioinf. 2005;61:900–906. doi: 10.1002/prot.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter CW, Jr, Duax WL. Did tRNA Synthetase Classes Arise on Opposite Strands of the Same Gene? Mol Cell. 2002;10:705–708. doi: 10.1016/s1097-2765(02)00688-3. [DOI] [PubMed] [Google Scholar]
- 49.Wills PR. The generation of meaningful information in molecular systems. Phil Trans R Soc A. 2016;A374:20150016. doi: 10.1098/rsta.20150066. [DOI] [PubMed] [Google Scholar]
- 50.Wills PR. Stepwise evolution of molecular biological coding. In: Pollack J, Bedau M, Husbands P, Ikegami T, Watson RA, editors. Artificial life IX. MIT Press; Cambridge: 2004. pp. 51–56. [Google Scholar]
- 51.Wills PR. Self-organization of genetic coding. J Theor Biol. 1993;162:267–287. doi: 10.1006/jtbi.1993.1087. [DOI] [PubMed] [Google Scholar]
- 52.Petrov AS, Williams LD. The Ancient Heart of the Ribosomal Large Subunit: A Response to Caetano-Anolles. J Mol Evol. 2015;80:166–170. doi: 10.1007/s00239-015-9678-8. [DOI] [PubMed] [Google Scholar]
- 53.Caetano-Anollés G, Wang M, Caetano-Anollés D. Structural Phylogenomics Retrodicts the Origin of the Genetic Code and Uncovers the Evolutionary Impact of Protein Flexibility. Plos One. 2013;8(8):e72225. doi: 10.1371/journal.pone.0072225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harish A, Caetano-Anollés G. Ribosomal History Reveals Origins of Modern Protein Synthesis. PLoS ONE. 2012;7(3):e32776. doi: 10.1371/journal.pone.0032776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfenden R, Lewis CA, Yuan Y, Carter CW., Jr Temperature dependence of amino acid hydrophobicities. Proc Nat Acad Sci USA. 2015;112(24):7484–7488. doi: 10.1073/pnas.1507565112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fersht AR. Transition-state structure as a unifying basis in protein-folding mechanisms: Contact order, chain topology, stability, and the extended nucleus mechanism. Proc Nat Acad Sci USA. 2000 Feb 15;97(4):1525–1529. doi: 10.1073/pnas.97.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dill KA, MacCallum JL. The Protein-Folding Problem, 50 Years On. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 58.Baker D. A surprising simplicity to protein folding. Nature. 2000 May;405(4):39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
- 59.Johnson Br, Lam SK. Self-organization, Natural Selection,and Evolution: Cellular Hardwareand Genetic Software. BioScience. 2010;60:879–885. doi: 10.1525/bio.2010.60.11.4. [DOI] [Google Scholar]
- 60.Füchslin RM, McCaskill JS. Evolutionary self-organization of cell-free genetic coding. Proc Natl Acad Sci USA. 2001;98:9185–9190. doi: 10.1073/pnas.151253198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Küippers B. Towards an Experimental Analysis of Molecular Self-Organization and Precellular Darwinian Evolution. Naturwissenschaften. 1979;66:228–243. doi: 10.1007/BF00571603. [DOI] [PubMed] [Google Scholar]
- 62.Eigen M, Schuster P. The Hypercyde: A Principle of Natural Self-Organization Part A: Emergence of the Hypercycle. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 63.Dennett DC. Darwin’s Dangerous Idea: Evolution and the Meanings of Life. Simon and Schuster; New York: 1995. [Google Scholar]
- 64.Sapienza PJ, Li L, Williams T, Lee AL, Carter CW., Jr An Ancestral Tryptophanyl-tRNA Synthetase Precursor Achieves High Catalytic Rate Enhancement without Ordered Ground-State Tertiary Structures. ACS Chemical Biology. 2016;11:1661–1668. doi: 10.1021/acschembio.5b01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carter J, Charles W, Chandrasekaran SN, Weinreb V, Li L, Williams T. Combining multi-mutant and modular thermodynamic cycles to measure energetic coupling networks in enzyme catalysis. In: Pearson A, Benedict J, editors. Structural Dynamics, American Crystallographic Association Annual Meeting. American Crystallographic Association; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caetano-Anollés G. Ancestral Insertions and Expansions of rRNA do not Support an Origin of the Ribosome in Its Peptidyl Transferase Center. J Mol Evol. 2015;80:162–165. doi: 10.1007/s00239-015-9677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun F-J, Caetano-Anollés G. Evolutionary Patterns in the Sequence and Structure of Transfer RNA: A Window into Early Translation and the Genetic Code. Plos One. 2008 Jul;3(7):e2799. doi: 10.1371/journal.pone.0002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pham Y, Li L, Kim A, Weinreb V, Butterfoss G, Kuhlman B, Carter CW., Jr A Minimal TrpRS Catalytic Domain Supports Sense/Antisense Ancestry of Class I and II Aminoacyl-tRNA Synthetases. Mol Cell. 2007;25:851–862. doi: 10.1016/j.molcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 69.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A Large Scale Test of Computational Protein Design: Folding and Stability of Nine Completely Redesigned Globular Proteins. J Mol Biol. 2003;332(2):449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 70.Chuang W-J, Abeygunawardana C, Pedersen PL, Mildvan AS. Two-Dimensional NMR, Circular Dichroism, and Fluorescence Studies of PP-50, a Synthetic ATP-Binding Peptide from the b-Subunit of Mitochondrial ATP Synthase. Biochem. 1992;31:7915–7921. doi: 10.1021/bi00149a024. [DOI] [PubMed] [Google Scholar]
- 71.Chuang W-J, Abeygunawardana C, Gittis AG, Pedersen PL, Mildvan AS. Solution Structure and Function in Trifluoroethanol of PP-50, an ATP-Binding Peptide from F1ATPase. Arch Biochem Biophys. 1992 May 1 10;319:110–122. doi: 10.1006/abbi.1995.1272. [DOI] [PubMed] [Google Scholar]
- 72.Mullen GP, Vaughn JB, Jr, Mildvan AS. Sequential Proton NMR Resonance Assignments, Circular Dichroism, and Structural Properties of a 50-Residue Substrate-Binding Peptide from DNA Polymerase I. Arch Biochem Biophys. 1993 Feb 1 15;301:174–183. doi: 10.1006/abbi.1993.1130. [DOI] [PubMed] [Google Scholar]
- 73.Fry DC, Byler DM, Sisu H, Brown EM, Kuby SA, Mildvan AS. Solution Structure of the 45-Residue MgATP-Binding Peptide of Adenylate Kinase As Examined by 2-D NMR, FTIR, and CD Spectroscopy. Biochem. 1988;27:3588–3598. doi: 10.1021/bi00410a009. [DOI] [PubMed] [Google Scholar]
- 74.Fry DC, Kuby SA, Mildvan AS. NMR Studies of the MgATP Binding Site of Adenylate Kinase and of a 45-Residue Peptide Fragment of the Enzyme. Biochem. 1985;24:4680–4694. doi: 10.1021/bi00338a030. [DOI] [PubMed] [Google Scholar]
- 75.Burbaum J, Schimmel P. Structural Relationships and the Classification of Aminoacyl-tRNA Synthetases. The Journal of Biological Chemistry. 1991;266(26):16965–16968. [PubMed] [Google Scholar]
- 76.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GLE, Hartley BS. Active Site Titration and Aminoacyl Adenylate Binding Stoichiometry of Amionacyl-tRNA Synthetases. Biochem. 1975;14(1):1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 77.Francklyn CS, First EA, Perona JJ, Hou Y-M. Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods. 2008;44:100–118. doi: 10.1016/j.ymeth.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirby AJ, Younas M. The reactivity of phosphate esters. Reactions of diesters with nucleophiles. Journal of the Chemical Society B. 1970;418:1165–1172. [Google Scholar]
- 79.Wolfenden R, Liang Y-L. Contributions of Solvent Water to Biological Group-Transfer Potentials: Mixed Anyhydrides of Phosphoric and Carboxylic Acids. Bioorganic Chemistry. 1989;17:486–489. [Google Scholar]
- 80.Schroeder GK, Wolfenden R. The Rate Enhancement Produced by the Ribosome: An Improved Model. Biochem. 2007;46:4037–4044. doi: 10.1021/bi602600p. [DOI] [PubMed] [Google Scholar]
- 81.Sievers A, Beringer M, Rodnina MV, Wolfenden R. The ribosome as an entropy trap. Proc Nat Acad Sci USA. 2004;101:7897–7901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shepherd J, Ibba M. Relaxed Substrate Specificity Leads to Extensive tRNA Mischarging by Streptococcus pneumoniae Class I and Class II Aminoacyl-tRNA Synthetases. mBio. 2014;5(5):e01656–01614. doi: 10.1128/mBio.01656-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ibba M, Soll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes and Development. 2004;18:731–738. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 84.Uter NT, Gruic-Sovulj I, Perona JJ. Amino Acid-dependent Transfer RNA Affinity in a Class I Aminoacyl-tRNA Synthetase. J Biol Chem. 2005 Jun 24;280(25):23966–23977. doi: 10.1074/jbc.M414259200. [DOI] [PubMed] [Google Scholar]
- 85.Sherlin LD, Perona JJ. tRNA-Dependent Active Site Assembly in a Class I Aminoacyl-tRNA Synthetase. Structure. 2003 May;11:591–603. doi: 10.1016/S0969-2126(03)00074-1. [DOI] [PubMed] [Google Scholar]
- 86.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Gen. 2007 Sep;8:675. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet. 2004 May;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 89.Carter CW, Jr, Chandrasekaran SN, Weinreb V, Li L, Williams T. Combining multi-mutant and modular thermodynamic cycles to measure energetic coupling networks in enzyme catalysis. Structural Dynamics. 2017;4:032101. doi: 10.1063/1.4974218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chandrasekaran SN, Carter CWJ. Adding torsional interaction terms to the Anisotropic Network Model improves the PATH performance, enabling detailed comparison with experimental rate data. Structural Dynamics. 2017;4:032103. doi: 10.1063/1.4976142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chandrasekaran SN, Das J, Dokholyan NV, Carter CW., Jr A modified PATH algorithm rapidly generates transition states comparable to those found by other well established algorithms. Structural Dynamics. 2016;3:012101. doi: 10.1063/1.4941599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weinreb V, Carter CW., Jr Mg2+-free B. stearothermophilus Tryptophanyl-tRNA Synthetase Activates Tryptophan With a Major Fraction of the Overall Rate Enhancement. Journal of the American Chemical Society. 2008;130:1488–1494. doi: 10.1021/ja076557x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weinreb V, Li L, Carter CW., Jr A Master Switch Couples Mg2+-Assisted Catalysis to Domain Motion in B. stearothermophilus Tryptophanyl-tRNA Synthetase. Structure. 2012;20 doi: 10.1016/j.str.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kapustina M, Weinreb V, Li L, Kuhlman B, Carter CW., Jr A Conformational Transition State Accompanies Tryptophan Activation by B. stearothermphilus Tryptophanyl-tRNA Synthetase. Structure. 2007;15:1272–1284. doi: 10.1016/j.str.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carter CW., Jr High-Dimensional Mutant and Modular Thermodynamic Cycles, Molecular Switching, and Free Energy Transduction. Annual Review of Biophysics. 2017;46:433–453. doi: 10.1146/annurev-biophys-070816-033811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Augustine J, Francklyn C. Design of an Active Fragment of a Class II Aminoacyl-tRNA Synthetase and Its Significance for Synthetase Evolution. Biochem. 1997;36:3473–3482. doi: 10.1021/bi962395y. [DOI] [PubMed] [Google Scholar]
- 97.Crick FHC. Codon-Anticodon Pairing: The Wobble Hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 98.Benner SA, Sassi SO, Gaucher EA. Molecular Paleoscience: Systems Biology from the Past. Advances in Enzymology and Related Areas of Molecular Biology. 2007;75:9–140. doi: 10.1002/9780471224464.ch1. [DOI] [PubMed] [Google Scholar]
- 99.Stackhouse J, Presnell SR, McGeehan GM, Nambiar KP, Benner SA. The Ribonuclease from an extinct bovid ruminant. FEBS Letters. 1990;262(1):104–106. doi: 10.1016/0014-5793(90)80164-e. [DOI] [PubMed] [Google Scholar]
- 100.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008 Feb;451(7):704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 101.Hanson-Smith V, Kolaczkowski B, Thornton JW. Robustness of Ancestral Sequence Reconstruction to Phylogenetic Uncertainty. Mol Biol Evol. 2010;27(9):1988–1999. doi: 10.1093/molbev/msq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 103.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bridgham JT, Carroll SM, Thornton JW. Evolution of Hormone-Receptor Complexity by Molecular Exploitation. Science. 2006 Apr;312(7):97–101. doi: 10.1126/science.1123348. 2006. [DOI] [PubMed] [Google Scholar]
- 105.Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- 106.Markowitz S, Drummond A, Nieselt K, Wills PR. Simulation model of prebiotic evolution of genetic coding. In: Rocha LM, Yaeger LS, Bedau MA, Floreano D, Goldstone RL, Vespignani A, editors. Artificial Life. Vol. 10. MIT Press; Cambridge, MA: 2006. pp. 152–157. [Google Scholar]
- 107.Wills PR, Nieselt K, McCaskill JS. Emergence of Coding and its Specificity as a Physico-Informatic Problem. Orig Life Evol Biosph. 2015 doi: 10.1007/s11084-015-9434-5. published online; pagination not yet available. [DOI] [PubMed] [Google Scholar]
- 108.Freeland SJ, Hurst LD. The Genetic Code is One in a Million. J Mol Evol. 1998;47:238–248. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- 109.Pervushin K, Vamvaca K, Vogeli B, Hilvert D. Structure and dynamics of a molten globular enzyme. Nat Struct Mol Biol. 2007 Dec;14:1202–1206. doi: 10.1038/nsmb1325. [DOI] [PubMed] [Google Scholar]
- 110.Hu H. Wild-type and molten globular chorismate mutase achieve comparable catalytic rates using very different enthalpy/entropy compensations. Science China. 2014;57(1):156–164. doi: 10.1007/s11426-013-5021-7. [DOI] [Google Scholar]
- 111.Patel SC, Bradley LH, Jinadasa SP, Hecht MH. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Prot Sci. 2009;18:1388–1400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moffet DA, Foley J, Hecht MH. Midpoint reduction potentials and heme binding stoichiometries of de novo proteins from designed combinatorial libraries. Biophys Chem. 2003;105:231–239. doi: 10.1016/s0301-4622(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 113.Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein Design by Binary Patterning of Polar and Non-polar Amino Acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 114.Safro M, Klipcan L. The mechanistic and evolutionary aspects of the 2′- and 3′-OH paradigm in biosynthetic machinery. Biology Direct. 2013;8:17. doi: 10.1186/1745-6150-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Klipcan L, Safro M. Amino acid biogenesis, evolution of the genetic code and aminoacyl-tRNA synthetases. Journal of Theoretical Biology. 2004;228:389–396. doi: 10.1016/j.jtbi.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 116.Smith TF, Hartman H. The evolution of Class II Aminoacyl-tRNA synthetases and the first code. FEBS Letters. 2015 Nov 30;589(23):3499–3507. doi: 10.1016/j.febslet.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 117.Perona JJ, Gruic-Sovulj I. Synthetic and Editing Mechanisms of Aminoacyl-tRNA Synthetases. Topics in Current Chemistry. 2013 doi: 10.1007/128_2013_456. [DOI] [PubMed] [Google Scholar]
- 118.Zhang C-M, Perona JJ, Ryu K, Francklyn C, Hou Y-M. Distinct Kinetic Mechanisms of the Two Classes of Aminoacyl-tRNA Synthetases. J Mol Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 119.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993 Jul;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nature Chemical Biology. 2013 Mar;9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo M, Yang X-L, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nature Reviews Molecular Cell Biology. 2010 Sep;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang XL, Schimmel P, Ewalt KL. Relationship of two human tRNA synthetases used in cell signaling. Trends in Biochem Sci. 2004;29(5):250–256. doi: 10.1016/j.tibs.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 123.Yang X-L, Guo M, Kapoor M, Ewalt KL, Otero FJ, Skene RJ, McRee DE, Schimmel P. Functional and Crystal Structure Analysis of Active Site Adaptations of a Potent Anti-Angiogenic Human tRNA Synthetase. Structure. 2007;15:793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Woese CR, Dugre DH, Saxinger WC, Dugre SA. The molecular basis for the genetic code. Proc Natl Acad Sci USA. 1966;55:966–974. doi: 10.1073/pnas.55.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wolfenden R. Experimental Measures of Amino Acid Hydrophobicity and the Topology of Transmembrane and Globular Proteins. Journal of General Physiology. 2007 May;129(5):357–362. doi: 10.1085/jgp.200709743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gibbs PR, Radzicka A, Wolfenden R. The Anomalous Hydrophilic Character of Proline. J Am Chem Soc. 1991;113:4714–4715. [Google Scholar]
- 127.Wolfenden R, Cullis PM, Southgate CCF. Water, Protein Folding, and the Genetic Code. Science. 1979;206:575–577. doi: 10.1126/science.493962. [DOI] [PubMed] [Google Scholar]
- 128.Wolfenden R, Andersson L, Cullis PM, Southgate CCF. Affinities of amino acid side chains for solvent water. Biochem. 1979;20:849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- 129.Radzicka A, Wolfenden R. Comparing the Polarities of the Amino Acids: Side-Chain Distribution Coefficients between the Vapor Phase, Cyclohexane, 1-0ctanol, and Neutral Aqueous Solution. Biochem. 1988;27(5):1664–1670. [Google Scholar]
- 130.Moelbert S, Emberly E, Tang C. Correlation between sequence hydrophobicity and surface-exposure pattern of database proteins. Prot Sci. 2004;13:752–762. doi: 10.1110/ps.03431704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Henderson BS, Schimmel P. RNA-RNA Interactions Between Oligonucleotide Substrates for Aminoacylation. Bioorganic & Medicinal Chemistry. 1997;5(6):1071–1079. doi: 10.1016/s0968-0896(97)00043-6. [DOI] [PubMed] [Google Scholar]
- 132.Francklyn C, Musier-Forsyth K, Schimmel P. Small RNA helices as substrates for aminoacylation and their relationship to charging of transfer RNAs. Euro J Biochem. 1992;206:315–321. doi: 10.1111/j.1432-1033.1992.tb16929.x. [DOI] [PubMed] [Google Scholar]
- 133.Francklyn C, Schimmel P. Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc Nati Acad Sci USA. 1990 Nov;87:8655–8659. doi: 10.1073/pnas.87.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Francklyn C, Schimmel P. Aminoacylation of RNA Minihelices with Alanine. Nature. 1989 Feb;337(2):478–481. doi: 10.1038/337478a0. 1989. [DOI] [PubMed] [Google Scholar]
- 135.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26(22):5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Carter CW., Jr Cradles for Molecular Evolution. New Scientist. 1975 Mar;27:784–787. [Google Scholar]
- 137.Carter CW, Jr, Kraut J. A Proposed Model for Interaction of Polypeptides with RNA. Proceedings of the National Academy of Sciences, USA. 1974;71(2):283–287. doi: 10.1073/pnas.71.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vestigian K, Woese CR, Goldenfeld N. Collective Evolution and the Genetic Code. Proc Nat Acad Sci USA. 2006;103:10696–10701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Woese C. Models for the Evolution of Codon Assignments. J Mol Biol. 1969;43:235–240. doi: 10.1016/0022-2836(69)90095-3. [DOI] [PubMed] [Google Scholar]
- 140.Woese C. The Genetic Code. Harper & Row; New York: 1967. [Google Scholar]
- 141.Leaver-Fay A, Jacak R, Stranges PB, Kuhlman B. A Generic Program for Multistate Protein Design. PLoS ONE. 2011;6(7):e20937. doi: 10.1371/journal.pone.0020937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.LéJohn HB, Cameron LE, Yang B, MacBeath G, Barker DS, Willams SA. Cloning and Analysis of a Constitutive Heat Shock (Cognate) Protein 70 Gene Inducible by L-Glutamine. J Biol Chem. 1994 Feb;269(11):4513–4522. [PubMed] [Google Scholar]
- 143.Yang B, LéJohn HB. NADP+-activable, NAD+-specific Glutamate Dehydrogenase Purification and Immunological Analysis. J Biol Chem. 1994 Feb;269(11):4506–4512. [PubMed] [Google Scholar]
- 144.LéJohn HB, Cameron LE, Yang B, Rennie SL. Molecular Characterization of an NAD-specific Glutamate Dehydrogenase Gene Inducible by L-Glutamine: Antisense Gene Pair Arrangement with L-Glutamine-Inducible Heat Shock 70-Like Protein Gene. J Biol Chem. 1994 Feb;269(11):4523–4531. [PubMed] [Google Scholar]
- 145.Linderstrøm-Lang KU. The Lane Medical Lectures. Stanford University Press; Stanford, CA: 1952. [Google Scholar]
- 146.Zull JE, Smith SK. Is genetic code redundancy related to retention of structural information in both DNA strands? TIBS. 1990;15:257–261. doi: 10.1016/0968-0004(90)90048-g. [DOI] [PubMed] [Google Scholar]
- 147.Yarus M. Life from an RNA World: The ancestor within. Harvard University Press; Cambridge, MA: 2011. [Google Scholar]
- 148.Yarus M, Widmann J, Knight R. RNA-amino acid binding: A stereochemical era for the genetic code. J Mol Evol. 2009;69:406–429. doi: 10.1007/s00239-009-9270-1. [DOI] [PubMed] [Google Scholar]
- 149.Koonin EV. The Logic of Chance: The Nature and Origin of Biological Evolution. Pearson Education; FT Press Science; Upper Saddle River, NJ: 2011. [Google Scholar]
- 150.Benner SA, Ellington AD, Tauer A. Modern metabolism as a palimpsest of the RNA world. Proc Nati Acad Sci USA. 1989 Sep;86:7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Danchin A. Archives or Palimpsests? Bacterial Genomes Unveil a Scenario for the Origin of Life. Biological Theory. 2007;2(1):1–10. [Google Scholar]
- 152.Fersht AR. Structure and Mechanism in Protein Science. W. H. Freeman and Company; New York: 1999. [Google Scholar]
- 153.Li R, Macnamara LM, Leuchter JD, Alexander RW, Cho SS. MD Simulations of tRNA and Aminoacyl-tRNA Synthetases: Dynamics, Folding, Binding, and Allostery. Int J Mol Sci. 2015;16:15872–15902. doi: 10.3390/ijms160715872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Budiman M, Knaggs MH, Fetrow JS, Alexander RW. Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase. PROTEINS, Structure, Function and Bioinformatics. 2007;68:670–689. doi: 10.1002/prot.21426. [DOI] [PubMed] [Google Scholar]
- 155.Kapustina M, Hermans J, Carter CW., Jr Potential of mean force estimation of the relative magnitude of the effect of errors in molecular mechanics approximations. Journal of Molecular Biology. 2006;362:1177–1180. [Google Scholar]
- 156.Kapustina M, Carter CW., Jr Computational Studies of Tryptophanyl-tRNA Synthetase Ligand Binding and Conformational Stability. J Mol Biol. 2006;362:1159–1180. doi: 10.1016/j.jmb.2006.06.078. [DOI] [PubMed] [Google Scholar]
- 157.Weinreb V, Li L, Kaguni LS, Campbell CL, Carter CW., Jr Mg2+-Assisted Catalysis by B. stearothermophilus TrpRS is Promoted by Allosteric Effects. Structure. 2009 Jul;17(15):1–13. doi: 10.1016/j.str.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wolf YI, Koonin EV. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biology Direct. 2007;2:14. doi: 10.1186/1745-6150-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Aravind L, Anantharaman V, Koonin EV. Monophyly of Class I Aminoacyl tRNASynthetase, USPA, ETFP, Photolyase, and PP-ATPase Nucleotide-Binding Domains: Implication for Protein Evolution in the RNAWorld. PROTEINS: Struct Funct Gen. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 160.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and Evolution of P-loop GTPases and Related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 161.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of Aminoacyl-tRNA Synthetases—Analysis of Unique Domain Architectures and Phylogenetic Trees Reveals a Complex History of Horizontal Gene Transfer Events. Genome Research. 1999;9:689–710. [PubMed] [Google Scholar]
- 162.Aravind L, Leipe DD, Koonin EV. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26(18):4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Silvert M, Simonson T. Creation and analysis of an algorithm creating overlapping genes. Laboratoire de Biochimie – École Polytechnique; Palaiseau, France: 2016. [Google Scholar]
- 164.Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Nat Acad Sci USA. 1963;49:517–521. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Eigen M. Selforganization of Matter and the Evolution of Biological Macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 166.Eigen M, McCaskill JS, Schuster P. Molecular Quasi-Species. J Phys Chem. 1988;92:6881–6891. [Google Scholar]
- 167.Bowman JC, Hud NV, Williams LD. The Ribosome Challenge to the RNA World. J Mol Evol. 2015;80:143–161. doi: 10.1007/s00239-015-9669-9. [DOI] [PubMed] [Google Scholar]
- 168.Petrov Anton S, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD. History of the ribosome and the origin of translation. PNAS. 2015 Dec 15;112(50):15396–15401. doi: 10.1073/pnas.1509761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hol WJG, van Duijnen PT, Berensen HJC. The α-helix dipole and the properties of proteins. Nature. 1978;273:443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- 170.Amyes TL, Richard JP. Specificity in Transition State Binding: The Pauling Model Revisited. Biochem. 2013;52:2021–2035. doi: 10.1021/bi301491r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Koonin EV, Novozhilov AS. Origin and Evolution of the Genetic Code: The Universal Enigma. IUBMB Life. 2009 Feb;61(2):99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Caetano-Anolles G, Kim HS, Mittenthal JE. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Nat Acad Sci USA. 2007 May 29;104(22):9358–9363. doi: 10.1073/pnas.0701214104. doi:10.1073pnas.0701214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Gen. 2015 Aug;16:472. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 174.Caetano-Anollés D, Caetano-Anollés G. Piecemeal Buildup of the Genetic Code, Ribosomes, and Genomes from Primordial tRNA Building Blocks. Life. 2016;6:43. doi: 10.3390/life6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Niwa N, Yamagishi Y, Murakami H, Suga H. A flexizyme that selectively charges amino acids activated by a water-friendly leaving group. Bioorg Med Chem Lett. 2009;19:3892–3894. doi: 10.1016/j.bmcl.2009.03.114. [DOI] [PubMed] [Google Scholar]
- 176.Suga H, Lohse PA, Szostak JW. Structural and Kinetic Characterization of an Acyl Transferase Ribozyme. J Am Chem Soc. 1998;120:1151–1156. doi: 10.1021/ja972472s. [DOI] [PubMed] [Google Scholar]
- 177.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA Synthetases into Two Classes Based on Mutually Exclusive Sets of Sequence Motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 178.Ardell DH, Andersson SGE. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006;34(3):893–904. doi: 10.1093/nar/gkj449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hofstadter DR. Gödel, Escher, Bach: an eternal golden braid. Basic Books, Inc; New York: 1979. [Google Scholar]
- 180.Fournier GP, Alm EJ. Ancestral Reconstruction of a Pre-LUCA Aminoacyl-tRNA Synthetase Ancestor Supports the Late Addition of Trp to the Genetic Code. J Mol Evol. 2015;80:171–185. doi: 10.1007/s00239-015-9672-1. [DOI] [PubMed] [Google Scholar]
- 181.Fournier GP, Andam CP, Alm EJ, Gogarten JP. Molecular Evolution of Aminoacyl tRNA Synthetase Proteins in the Early History of Life. Orig Life Evol Biosph. 2011;41:621–632. doi: 10.1007/s11084-011-9261-2. [DOI] [PubMed] [Google Scholar]
- 182.SAS. JMP: The Statistical Discovery Software. V.10 edn. SAS Institute; Cary NC, Cary, NC: 2015. [Google Scholar]